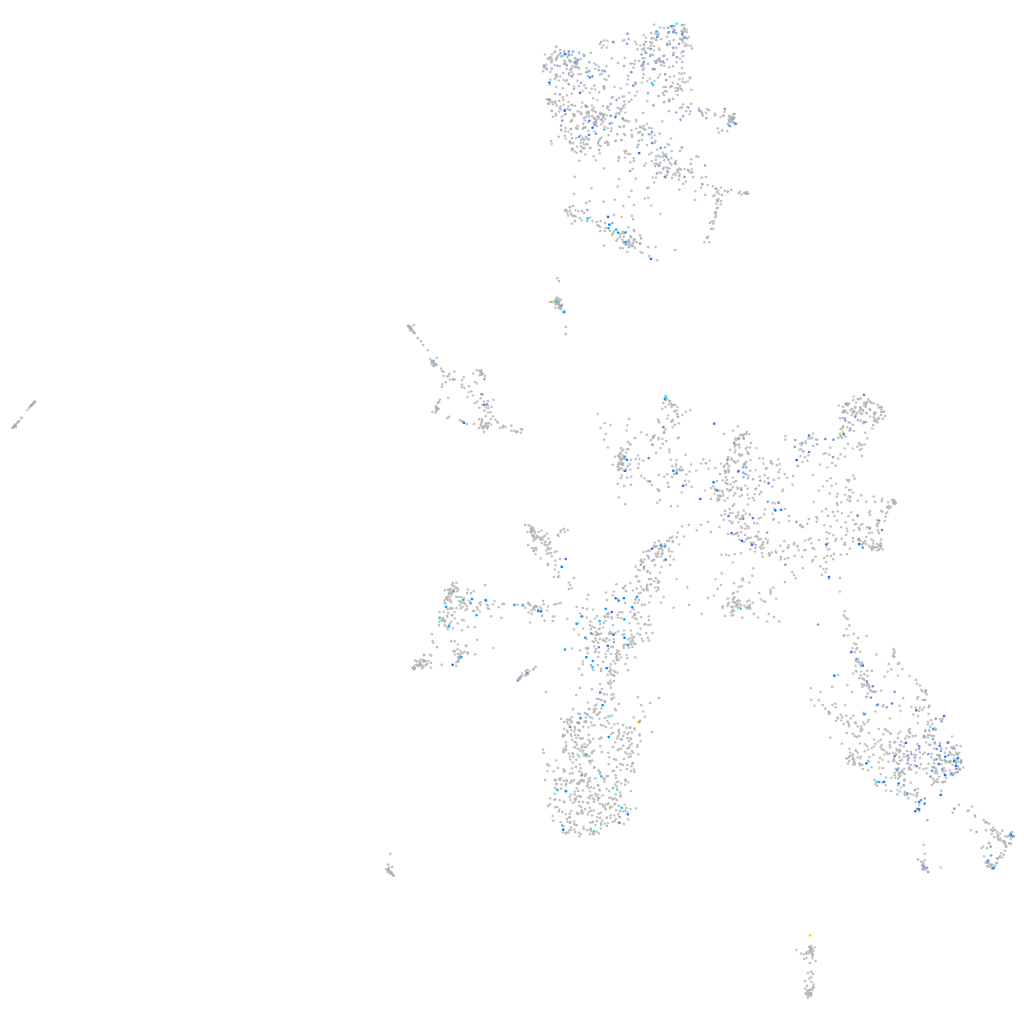
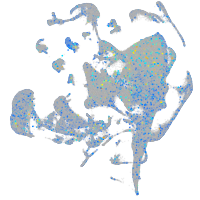
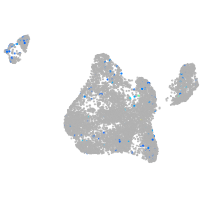
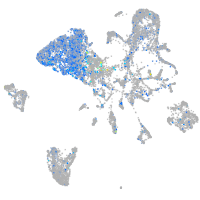
Ecm29 proteasome adaptor and scaffold
ZFIN 
Expression by stage/cluster


Correlated gene expression