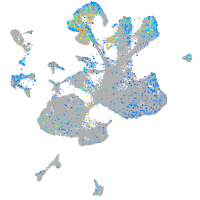
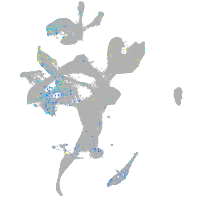
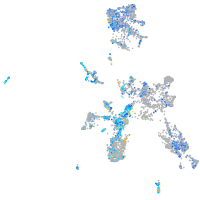
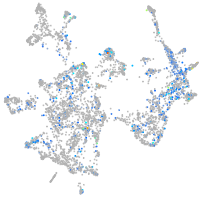
endothelin converting enzyme 2b
ZFIN 
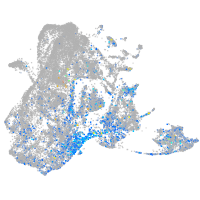
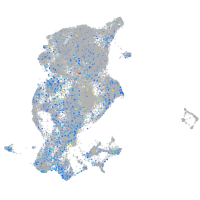
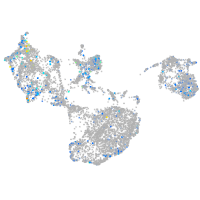
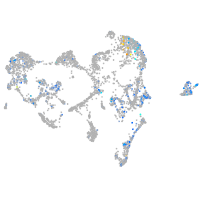
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl4 | 0.165 | hmgb2a | -0.123 |
| rtn1b | 0.163 | hmga1a | -0.078 |
| stmn2a | 0.161 | fabp7a | -0.071 |
| zgc:65894 | 0.157 | ahcy | -0.066 |
| elavl3 | 0.151 | hmgb2b | -0.066 |
| ywhag2 | 0.151 | rrm1 | -0.065 |
| sv2a | 0.149 | pcna | -0.064 |
| stx1b | 0.146 | stmn1a | -0.062 |
| stxbp1a | 0.144 | msi1 | -0.062 |
| syt1a | 0.144 | chaf1a | -0.062 |
| snap25a | 0.143 | rplp0 | -0.062 |
| gabrb4 | 0.140 | lbr | -0.060 |
| sprn | 0.139 | ccnd1 | -0.059 |
| eno2 | 0.138 | dut | -0.059 |
| sncb | 0.138 | mki67 | -0.058 |
| celf4 | 0.138 | ccna2 | -0.058 |
| gng3 | 0.137 | rpsa | -0.058 |
| stmn1b | 0.137 | nutf2l | -0.058 |
| tuba2 | 0.134 | mcm7 | -0.058 |
| islr2 | 0.133 | mdka | -0.056 |
| rtn1a | 0.132 | dek | -0.055 |
| maptb | 0.132 | rps12 | -0.054 |
| scg2b | 0.131 | rpa3 | -0.054 |
| tkta | 0.131 | cks1b | -0.054 |
| gap43 | 0.130 | rrm2 | -0.054 |
| xpr1a | 0.126 | lig1 | -0.054 |
| myt1la | 0.126 | rps20 | -0.054 |
| cplx2 | 0.126 | zgc:110216 | -0.054 |
| celf5a | 0.123 | zgc:110425 | -0.053 |
| nrxn2a | 0.122 | cx43.4 | -0.053 |
| id4 | 0.121 | neurod4 | -0.052 |
| zgc:153426 | 0.121 | rps27a | -0.052 |
| syt2a | 0.120 | XLOC-042899 | -0.052 |
| ywhag1 | 0.120 | CABZ01005379.1 | -0.052 |
| pbx3b | 0.119 | ran | -0.052 |