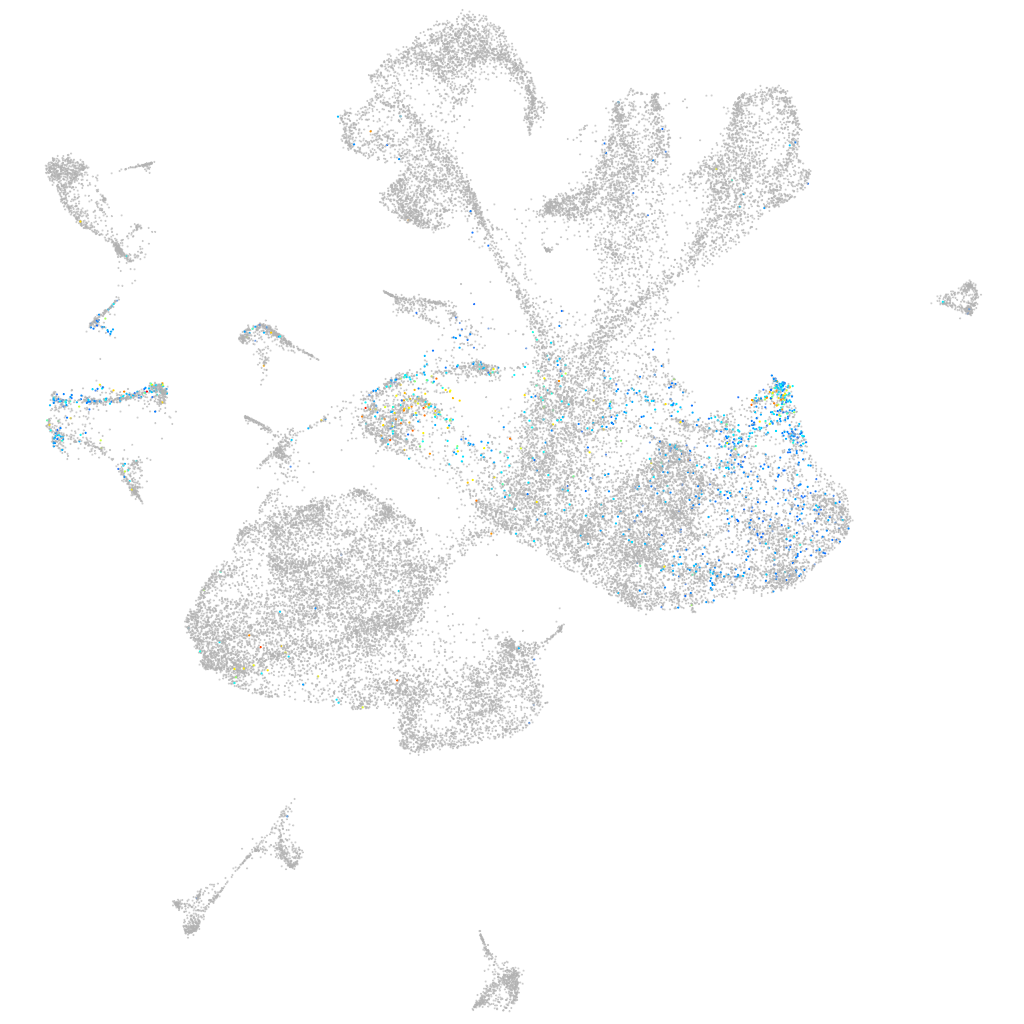
death domain containing 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkeyp-110a12.4 | 0.238 | tmsb4x | -0.118 |
| smkr1 | 0.234 | rtn1a | -0.096 |
| hoxb13a | 0.232 | marcksl1b | -0.087 |
| SPATA1 | 0.229 | stmn1b | -0.086 |
| krt18a.1 | 0.228 | ckbb | -0.085 |
| si:ch211-155m12.5 | 0.227 | fabp3 | -0.081 |
| si:dkeyp-69c1.9 | 0.226 | si:dkey-276j7.1 | -0.080 |
| tppp3 | 0.216 | gpm6ab | -0.078 |
| c8h22orf15 | 0.211 | tmsb | -0.078 |
| tbata | 0.208 | elavl3 | -0.077 |
| spata18 | 0.206 | ywhah | -0.075 |
| enkur | 0.205 | myt1b | -0.072 |
| si:ch211-163l21.7 | 0.200 | gng3 | -0.068 |
| hoxc13b | 0.198 | si:ch211-133n4.4 | -0.067 |
| si:ch211-250c4.5 | 0.193 | tmeff1b | -0.067 |
| eve1 | 0.192 | rnasekb | -0.064 |
| lmna | 0.190 | celf2 | -0.064 |
| cdkl1 | 0.188 | fez1 | -0.064 |
| si:ch73-265d7.2 | 0.186 | elavl4 | -0.063 |
| pacrg | 0.185 | stx1b | -0.062 |
| rab36 | 0.185 | stxbp1a | -0.062 |
| bicc2 | 0.184 | atp6v0cb | -0.061 |
| hoxa13a | 0.184 | nsg2 | -0.060 |
| meig1 | 0.183 | nova1 | -0.060 |
| lrrc6 | 0.181 | tuba1c | -0.060 |
| krt8 | 0.180 | sncb | -0.060 |
| daw1 | 0.179 | rtn1b | -0.059 |
| pkd1b | 0.178 | syt2a | -0.059 |
| spata4 | 0.177 | vamp2 | -0.059 |
| ak7a | 0.174 | mllt11 | -0.059 |
| dnali1 | 0.174 | hmgb3a | -0.059 |
| hoxa13b | 0.173 | tubb5 | -0.058 |
| ccdc169 | 0.172 | cplx2 | -0.058 |
| capsla | 0.172 | nova2 | -0.058 |
| zgc:55461 | 0.172 | si:dkeyp-75h12.5 | -0.057 |