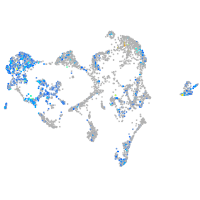
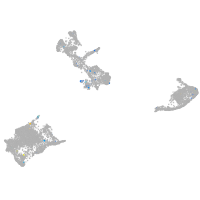
desmocollin 2 like
ZFIN 
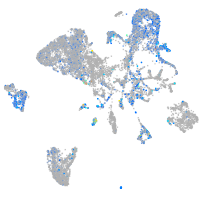
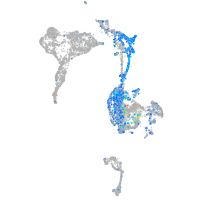
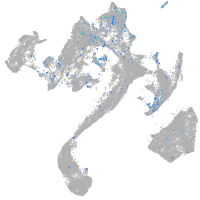
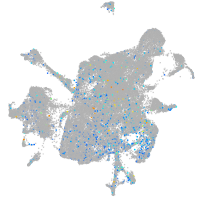
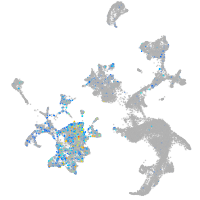
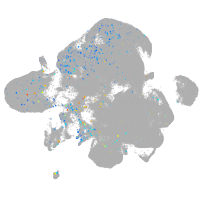
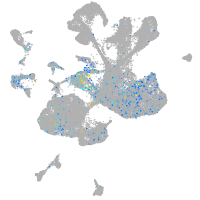
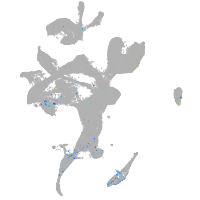
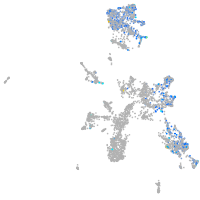
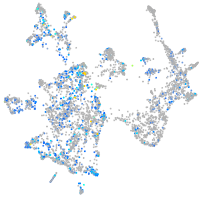
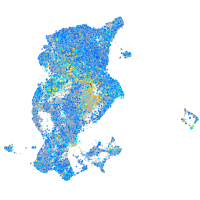
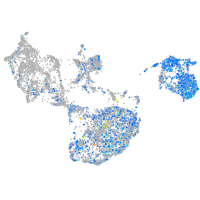
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108181079 | 0.671 | ppig | -0.209 |
| malt2 | 0.612 | kif23 | -0.206 |
| efr3ba | 0.580 | waca | -0.200 |
| emilin2a | 0.580 | ddx18 | -0.198 |
| kif3cb | 0.580 | hnrnpa1a | -0.198 |
| si:ch1073-513e17.1 | 0.580 | sec62 | -0.197 |
| si:dkey-1h24.2 | 0.580 | srrt | -0.192 |
| adcy7 | 0.569 | yipf5 | -0.189 |
| sema6a | 0.547 | mad2l1 | -0.189 |
| ano1 | 0.546 | rbm4.2 | -0.181 |
| ccdc96 | 0.536 | u2af2a | -0.180 |
| march8 | 0.535 | clptm1 | -0.179 |
| arhgap36 | 0.527 | gar1 | -0.178 |
| LO018166.1 | 0.523 | brd2a | -0.178 |
| cenpv | 0.522 | nucks1a | -0.176 |
| camkva | 0.521 | srrm2 | -0.175 |
| znf408 | 0.520 | rnf113a | -0.174 |
| ugt1b5 | 0.517 | nsrp1 | -0.174 |
| si:ch73-223p23.2 | 0.517 | sf3b2 | -0.174 |
| zic1 | 0.514 | pum3 | -0.174 |
| b3gnt2l | 0.513 | ncaph | -0.173 |
| pcdh18a | 0.513 | baz1b | -0.170 |
| trim62 | 0.513 | fam50a | -0.169 |
| CABZ01101984.1 | 0.507 | map7d3 | -0.166 |
| CABZ01065076.1 | 0.505 | aurkb | -0.166 |
| agmo | 0.505 | kri1 | -0.165 |
| DPYSL2 (1 of many) | 0.505 | plk1 | -0.165 |
| clrn2 | 0.495 | fnbp4 | -0.163 |
| zgc:63568 | 0.494 | eif4a1a | -0.162 |
| sept4b | 0.494 | ISCU | -0.159 |
| zgc:56585 | 0.492 | prkrip1 | -0.157 |
| hoxa13b | 0.487 | exosc10 | -0.157 |
| prdm2a | 0.485 | mad1l1 | -0.157 |
| BX957317.2 | 0.484 | etv5a | -0.157 |
| crispld1a | 0.483 | msl1b | -0.155 |