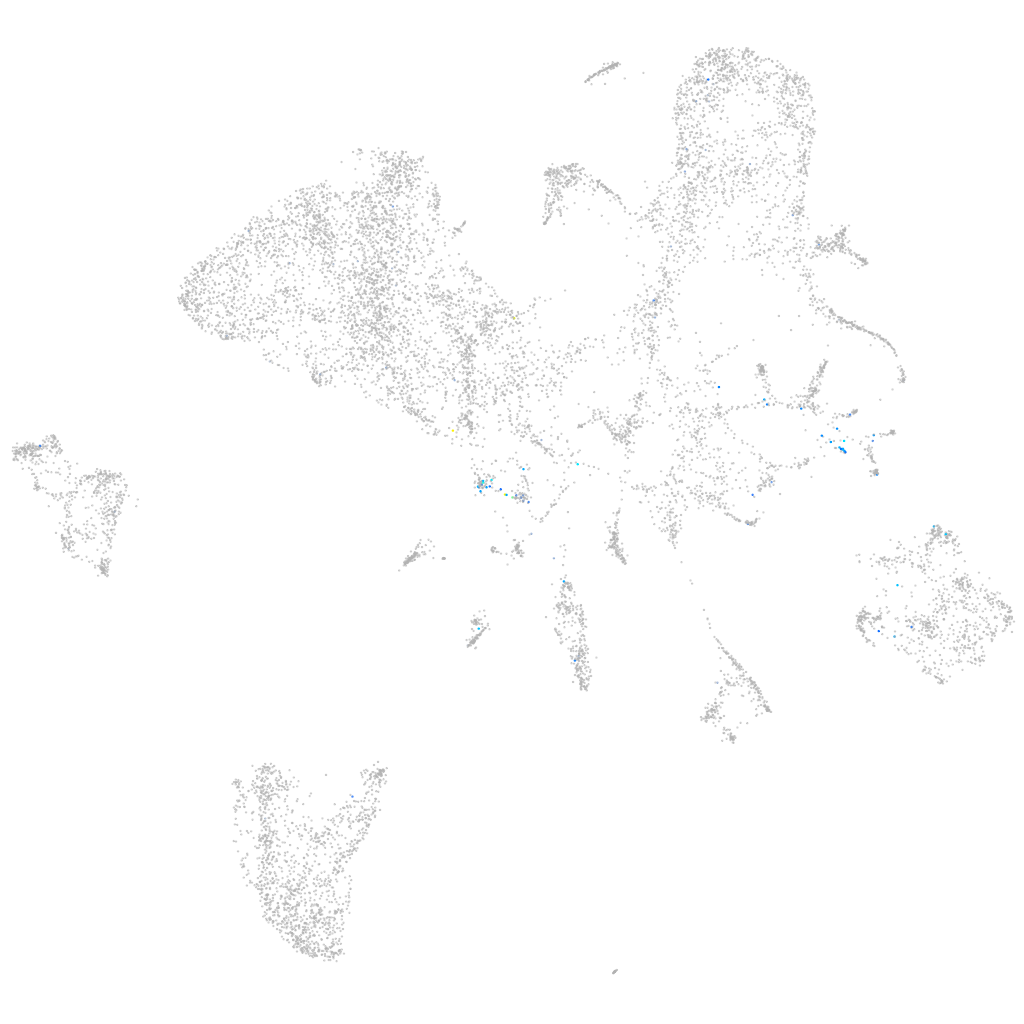
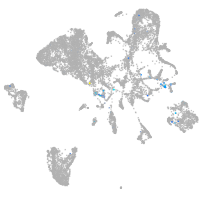
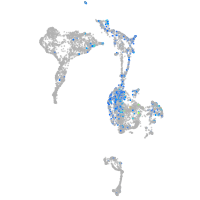
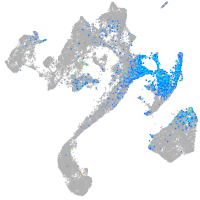
dorsal inhibitory axon guidance protein
ZFIN 
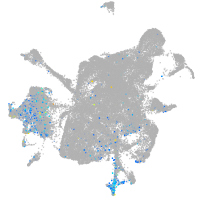
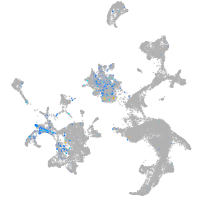
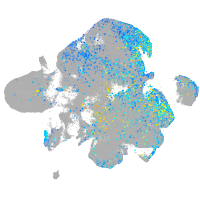
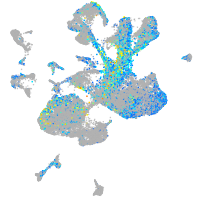
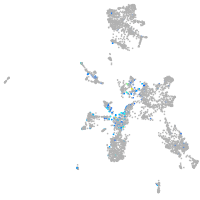
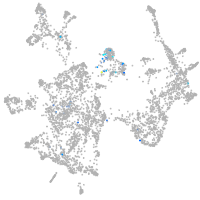
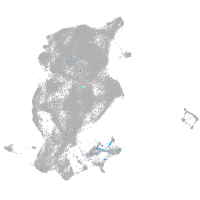
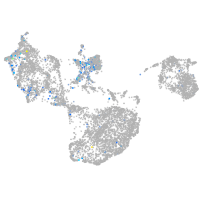
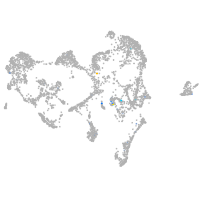
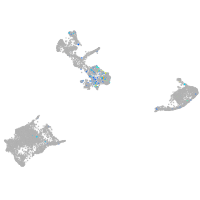
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nhlh2 | 0.239 | eef1da | -0.069 |
| nrcamb | 0.221 | aldob | -0.067 |
| slc17a7b | 0.217 | sod1 | -0.065 |
| ebf2 | 0.208 | gapdh | -0.062 |
| elavl3 | 0.201 | cebpd | -0.060 |
| inavaa | 0.199 | glud1b | -0.060 |
| LOC792894 | 0.197 | eno3 | -0.058 |
| lgi1a | 0.197 | hdlbpa | -0.058 |
| vim | 0.197 | nupr1b | -0.057 |
| scrt2 | 0.196 | gsta.1 | -0.055 |
| cnrip1a | 0.194 | gstr | -0.054 |
| frrs1l | 0.194 | cx32.3 | -0.054 |
| LOC103909675 | 0.192 | gamt | -0.053 |
| LOC110438274 | 0.190 | fbp1b | -0.053 |
| myt1b | 0.188 | rmdn1 | -0.052 |
| LOC101886459 | 0.187 | sod2 | -0.051 |
| LOC100537384 | 0.184 | pklr | -0.051 |
| si:dkey-183p4.10 | 0.184 | chchd10 | -0.050 |
| BX530077.1 | 0.183 | prdx6 | -0.050 |
| nova2 | 0.182 | ahcy | -0.050 |
| dlb | 0.181 | gstt1a | -0.050 |
| myt1a | 0.179 | gstp1 | -0.049 |
| tuba1c | 0.179 | ckba | -0.049 |
| si:dkey-187i8.2 | 0.177 | slc16a10 | -0.049 |
| tuba1a | 0.176 | dhdhl | -0.049 |
| prph2l | 0.176 | atp1b1a | -0.049 |
| hs3st3l | 0.173 | scp2a | -0.048 |
| neurod2 | 0.173 | BX908782.3 | -0.047 |
| ncam1a | 0.171 | atp5f1c | -0.047 |
| insm1a | 0.171 | agxta | -0.046 |
| zgc:112408 | 0.168 | nipsnap3a | -0.046 |
| jagn1a | 0.167 | aldh9a1a.1 | -0.046 |
| CABZ01079550.1 | 0.167 | gatm | -0.045 |
| sncb | 0.167 | dap | -0.045 |
| epb41a | 0.166 | mif | -0.045 |