2'-deoxynucleoside 5'-phosphate N-hydrolase 1
ZFIN 
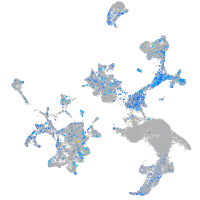
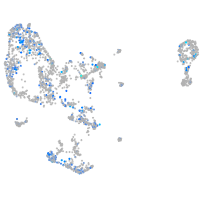
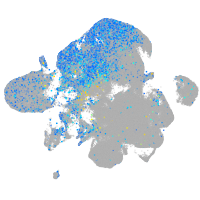
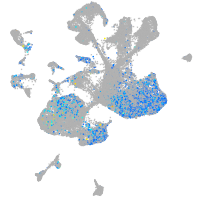
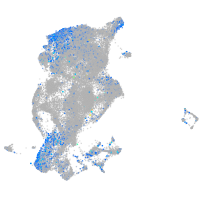
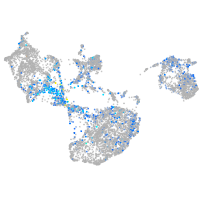
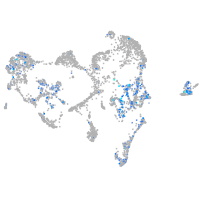
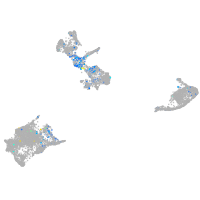
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.451 | h1f0 | -0.158 |
| pcna | 0.439 | atp2b1a | -0.144 |
| stmn1a | 0.417 | calm1a | -0.142 |
| rpa2 | 0.413 | tob1b | -0.141 |
| si:ch211-156b7.4 | 0.388 | rnasekb | -0.140 |
| rrm2 | 0.388 | atp1a3b | -0.140 |
| rrm1 | 0.387 | CR383676.1 | -0.138 |
| rpa3 | 0.386 | otofb | -0.137 |
| nasp | 0.384 | gpx2 | -0.136 |
| zgc:110540 | 0.375 | calml4a | -0.136 |
| fen1 | 0.368 | tspan13b | -0.135 |
| dhfr | 0.367 | nptna | -0.134 |
| si:dkey-6i22.5 | 0.358 | cd164l2 | -0.134 |
| chaf1a | 0.355 | tuba1a | -0.132 |
| banf1 | 0.347 | dnajc5b | -0.130 |
| mibp | 0.340 | osbpl1a | -0.130 |
| cks1b | 0.339 | pkig | -0.129 |
| tubb2b | 0.331 | rbm24a | -0.129 |
| dnajc9 | 0.330 | ckbb | -0.129 |
| unga | 0.329 | CR925719.1 | -0.128 |
| rnaseh2a | 0.328 | lrrc73 | -0.126 |
| shmt1 | 0.328 | prr15la | -0.125 |
| CABZ01005379.1 | 0.324 | kif1aa | -0.125 |
| slbp | 0.323 | slc17a8 | -0.124 |
| mcm7 | 0.322 | si:ch73-15n15.3 | -0.124 |
| ranbp1 | 0.321 | calm1b | -0.124 |
| hmgb2a | 0.320 | atp1b2b | -0.124 |
| dek | 0.318 | pcsk5a | -0.123 |
| orc4 | 0.318 | gipc3 | -0.123 |
| nutf2l | 0.315 | bdnf | -0.122 |
| lig1 | 0.314 | emb | -0.122 |
| rfc4 | 0.312 | myo6b | -0.122 |
| prim2 | 0.312 | nupr1a | -0.122 |
| anp32b | 0.311 | apba1a | -0.122 |
| mis12 | 0.311 | aldocb | -0.122 |