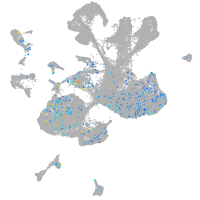
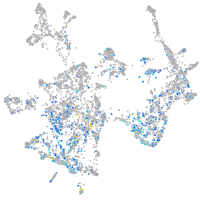
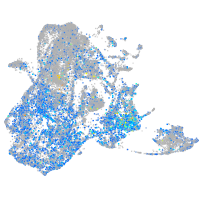
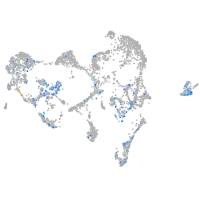
"DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1"
ZFIN 
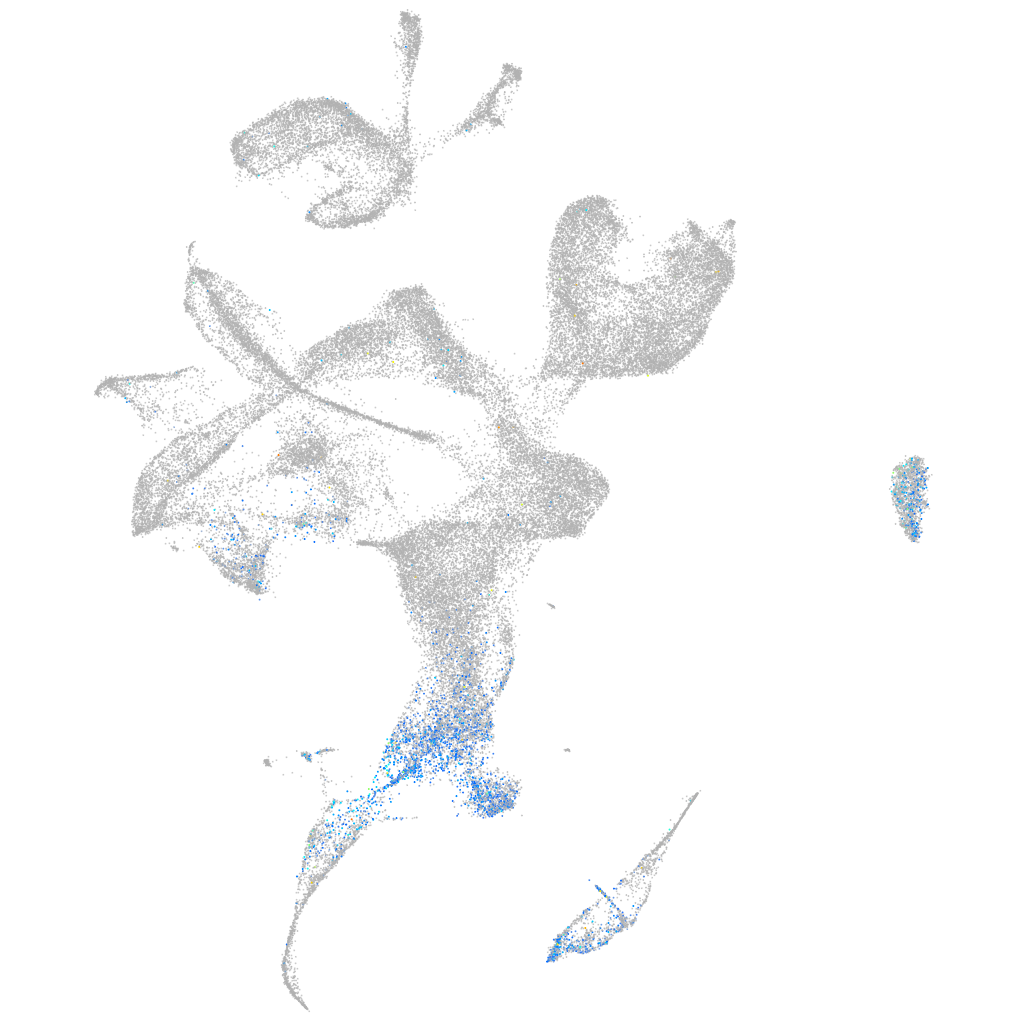
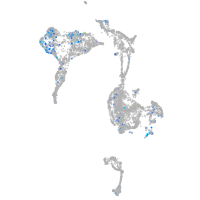
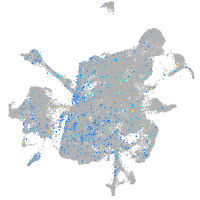
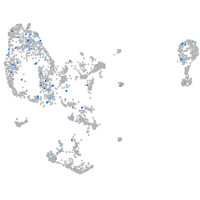
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.269 | ckbb | -0.188 |
| nop58 | 0.248 | ptmaa | -0.155 |
| dkc1 | 0.245 | gpm6ab | -0.152 |
| nop2 | 0.242 | ptmab | -0.152 |
| fabp11a | 0.239 | gpm6aa | -0.151 |
| si:dkey-102m7.3 | 0.237 | tuba1c | -0.144 |
| snu13b | 0.237 | rnasekb | -0.129 |
| col15a1b | 0.236 | hmgb1a | -0.124 |
| eif4ebp3l | 0.234 | atp6v0cb | -0.123 |
| fbl | 0.233 | hmgn6 | -0.123 |
| nhp2 | 0.233 | snap25b | -0.117 |
| rsl1d1 | 0.233 | CR383676.1 | -0.117 |
| nop56 | 0.232 | foxg1b | -0.115 |
| pprc1 | 0.230 | cadm3 | -0.109 |
| cdon | 0.229 | btg1 | -0.108 |
| gnl3 | 0.225 | stmn1b | -0.107 |
| rrp1 | 0.223 | epb41a | -0.106 |
| si:ch211-217k17.7 | 0.222 | rtn1a | -0.106 |
| paics | 0.222 | neurod4 | -0.103 |
| her9 | 0.219 | h3f3c | -0.103 |
| nop10 | 0.217 | ubc | -0.103 |
| nip7 | 0.213 | pvalb2 | -0.102 |
| prps1a | 0.212 | atpv0e2 | -0.101 |
| mybbp1a | 0.210 | atp6v1g1 | -0.100 |
| rcl1 | 0.209 | crx | -0.100 |
| twistnb | 0.209 | pvalb1 | -0.100 |
| cad | 0.208 | tuba1a | -0.099 |
| wdr43 | 0.207 | syt5b | -0.099 |
| CU929070.1 | 0.204 | atp6v1e1b | -0.099 |
| rpl7l1 | 0.203 | ndrg4 | -0.099 |
| ncl | 0.203 | marcksl1b | -0.098 |
| bms1 | 0.202 | marcksl1a | -0.096 |
| ppan | 0.201 | pdcl | -0.095 |
| ddx27 | 0.200 | cspg5a | -0.094 |
| ebna1bp2 | 0.199 | sypb | -0.094 |