DnaJ heat shock protein family (Hsp40) member B6b
ZFIN 
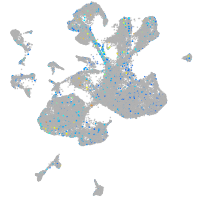
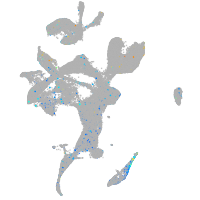
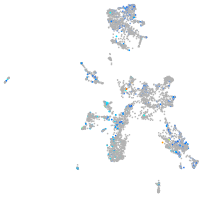
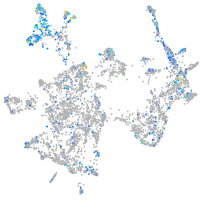
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mnx1 | 0.094 | rplp1 | -0.058 |
| lhx4 | 0.087 | stmn1a | -0.055 |
| mnx2b | 0.078 | hmgn2 | -0.051 |
| gapdhs | 0.073 | pcna | -0.051 |
| rnasekb | 0.072 | rplp2l | -0.051 |
| aldocb | 0.070 | si:dkey-151g10.6 | -0.051 |
| mnx2a | 0.070 | hmgb2b | -0.049 |
| ckbb | 0.068 | hmgb2a | -0.049 |
| atp6v1e1b | 0.067 | dut | -0.048 |
| tpi1b | 0.066 | her2 | -0.046 |
| anxa13 | 0.066 | lbr | -0.046 |
| ndrg3a | 0.063 | hmga1a | -0.045 |
| atp6v1g1 | 0.062 | rps20 | -0.044 |
| si:ch73-119p20.1 | 0.061 | rps29 | -0.043 |
| mllt11 | 0.061 | nasp | -0.042 |
| atp6v0cb | 0.061 | rpl35a | -0.042 |
| atpv0e2 | 0.060 | rpl37 | -0.042 |
| zgc:65894 | 0.060 | nutf2l | -0.041 |
| nptna | 0.060 | rpl39 | -0.041 |
| gdi1 | 0.060 | rpl26 | -0.040 |
| ywhag2 | 0.060 | rps19 | -0.040 |
| ache | 0.059 | her12 | -0.039 |
| mapkapk2b | 0.059 | chaf1a | -0.039 |
| emc7 | 0.059 | rps25 | -0.039 |
| abcg4a | 0.059 | mki67 | -0.039 |
| gpia | 0.059 | slc1a3a | -0.039 |
| slc18a3a | 0.058 | rps21 | -0.039 |
| ndufa4 | 0.057 | rps12 | -0.038 |
| calm3a | 0.057 | rps14 | -0.038 |
| jpt1b | 0.056 | dek | -0.038 |
| bsg | 0.055 | si:ch73-281n10.2 | -0.038 |
| fez1 | 0.055 | ccnd1 | -0.037 |
| pygmb | 0.055 | mibp | -0.037 |
| chga | 0.055 | rpl36 | -0.037 |
| gabarapb | 0.054 | rpl23 | -0.037 |