DnaJ heat shock protein family (Hsp40) member B5
ZFIN 
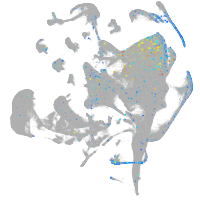
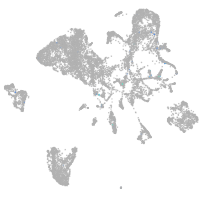
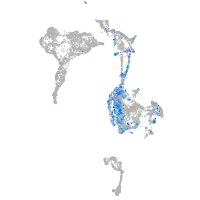
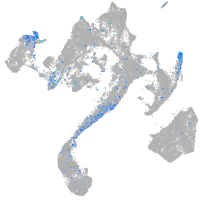
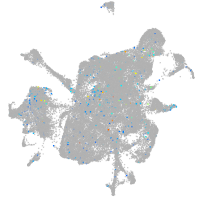
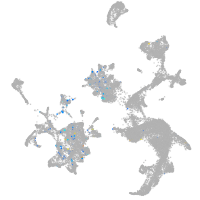
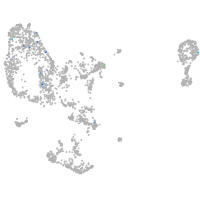
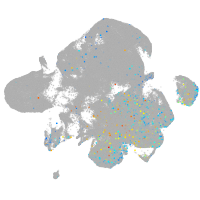
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn2a | 0.147 | XLOC-003690 | -0.094 |
| stx1b | 0.145 | id1 | -0.091 |
| stxbp1a | 0.144 | hmgb2a | -0.086 |
| ywhag2 | 0.141 | her15.1 | -0.085 |
| sncb | 0.140 | mdka | -0.081 |
| elavl4 | 0.139 | cldn5a | -0.077 |
| atp6v0cb | 0.136 | XLOC-003689 | -0.074 |
| snap25a | 0.135 | rplp1 | -0.074 |
| zgc:65894 | 0.134 | sox3 | -0.073 |
| atp1a3a | 0.133 | btg2 | -0.071 |
| cplx2l | 0.132 | sdc4 | -0.070 |
| gng3 | 0.131 | vamp3 | -0.070 |
| vamp2 | 0.131 | si:dkey-151g10.6 | -0.068 |
| sncgb | 0.130 | tspan7 | -0.068 |
| sypb | 0.130 | sparc | -0.068 |
| rbfox1 | 0.130 | notch3 | -0.066 |
| kcnc3a | 0.129 | rps20 | -0.065 |
| rtn1b | 0.129 | XLOC-003692 | -0.063 |
| cplx2 | 0.129 | her12 | -0.063 |
| calm1b | 0.125 | COX7A2 (1 of many) | -0.063 |
| syn2a | 0.124 | her4.2 | -0.062 |
| eno2 | 0.124 | rps2 | -0.062 |
| aplp1 | 0.124 | zgc:56493 | -0.061 |
| syt2a | 0.123 | sox19a | -0.061 |
| evlb | 0.122 | msi1 | -0.061 |
| cdk5r2a | 0.121 | rpl39 | -0.061 |
| tuba2 | 0.118 | her4.1 | -0.061 |
| map1aa | 0.118 | msna | -0.059 |
| ndufa4 | 0.116 | sb:cb81 | -0.059 |
| camk2d2 | 0.115 | gfap | -0.059 |
| tmem59l | 0.115 | hmgb2b | -0.059 |
| mllt11 | 0.115 | dut | -0.058 |
| sh3gl2a | 0.115 | stmn1a | -0.058 |
| ywhah | 0.114 | rpl12 | -0.057 |
| tmsb2 | 0.114 | rpl10a | -0.057 |