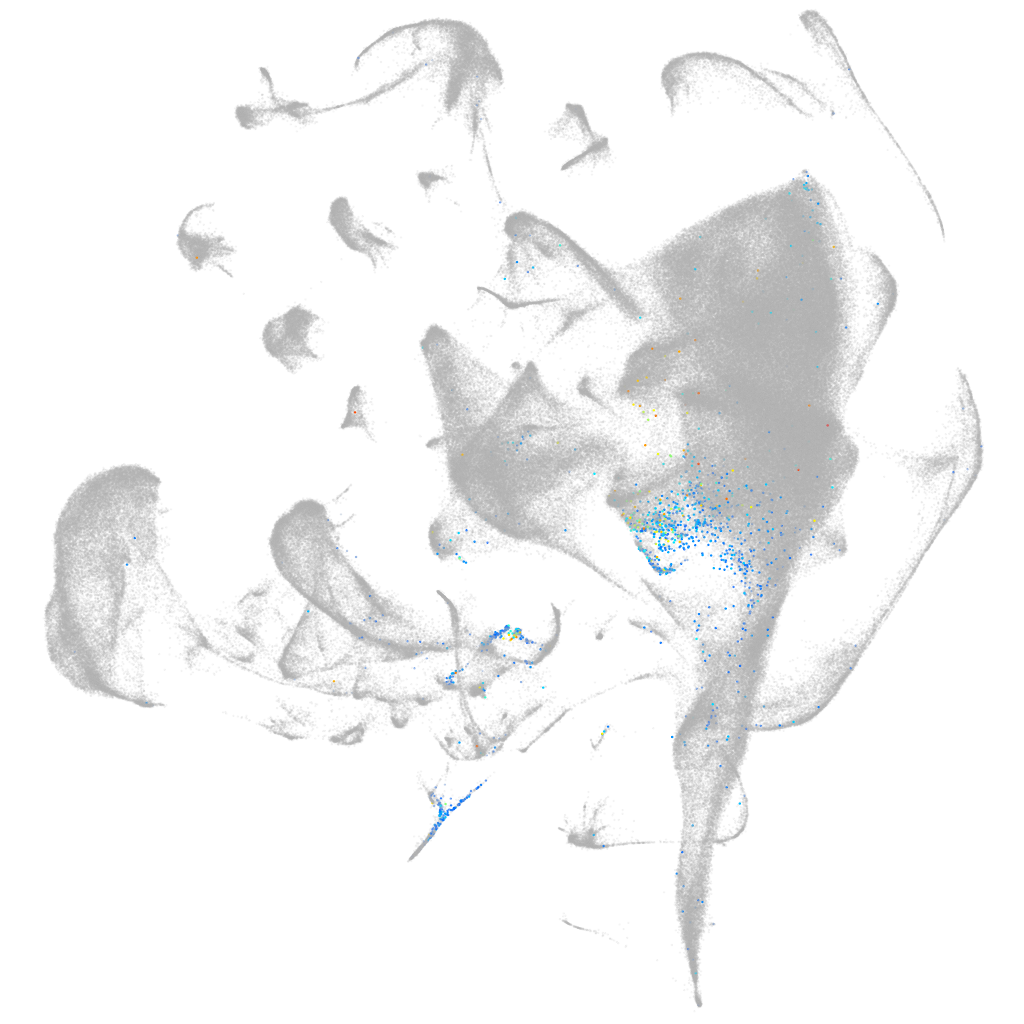
"dynein, axonemal, heavy chain 5"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.238 | stmn1b | -0.023 |
| foxj1a | 0.218 | gpm6ab | -0.022 |
| ccdc151 | 0.204 | celf2 | -0.021 |
| cfap126 | 0.201 | gng3 | -0.019 |
| enkur | 0.196 | pvalb2 | -0.019 |
| tbata | 0.195 | rtn1a | -0.018 |
| tekt2 | 0.195 | rtn1b | -0.018 |
| dnali1 | 0.192 | actc1b | -0.017 |
| si:ch211-163l21.7 | 0.192 | pvalb1 | -0.017 |
| morn3 | 0.190 | ywhag2 | -0.017 |
| tekt1 | 0.189 | cyt1 | -0.016 |
| si:dkey-27p23.3 | 0.186 | cyt1l | -0.016 |
| spag6 | 0.183 | elavl4 | -0.016 |
| pacrg | 0.182 | fxyd6l | -0.016 |
| spaca9 | 0.182 | krt4 | -0.016 |
| ttc25 | 0.182 | mab21l1 | -0.016 |
| ak7a | 0.180 | ndrg4 | -0.016 |
| si:ch211-155m12.5 | 0.180 | nsg2 | -0.016 |
| ccdc114 | 0.179 | snap25b | -0.016 |
| tekt4 | 0.179 | sypb | -0.016 |
| ak9 | 0.179 | arhgdig | -0.015 |
| ccdc173 | 0.178 | atp1a3a | -0.015 |
| ccdc169 | 0.177 | icn2 | -0.015 |
| cfap52 | 0.176 | krtt1c19e | -0.015 |
| gb:eh456644 | 0.176 | olfm1b | -0.015 |
| iqcg | 0.175 | rbfox1 | -0.015 |
| nme8 | 0.175 | si:ch211-105c13.3 | -0.015 |
| si:ch211-226m7.4 | 0.172 | si:dkey-276j7.1 | -0.015 |
| dnajc5b | 0.169 | stmn2a | -0.015 |
| efhc2 | 0.169 | stx1b | -0.015 |
| nme5 | 0.169 | tmsb1 | -0.015 |
| ribc2 | 0.168 | ywhah | -0.015 |
| ccdc65 | 0.167 | col1a2 | -0.014 |
| dnaaf3 | 0.167 | elavl3 | -0.014 |
| smkr1 | 0.167 | gap43 | -0.014 |