"dynein, axonemal, assembly factor 3"
ZFIN 
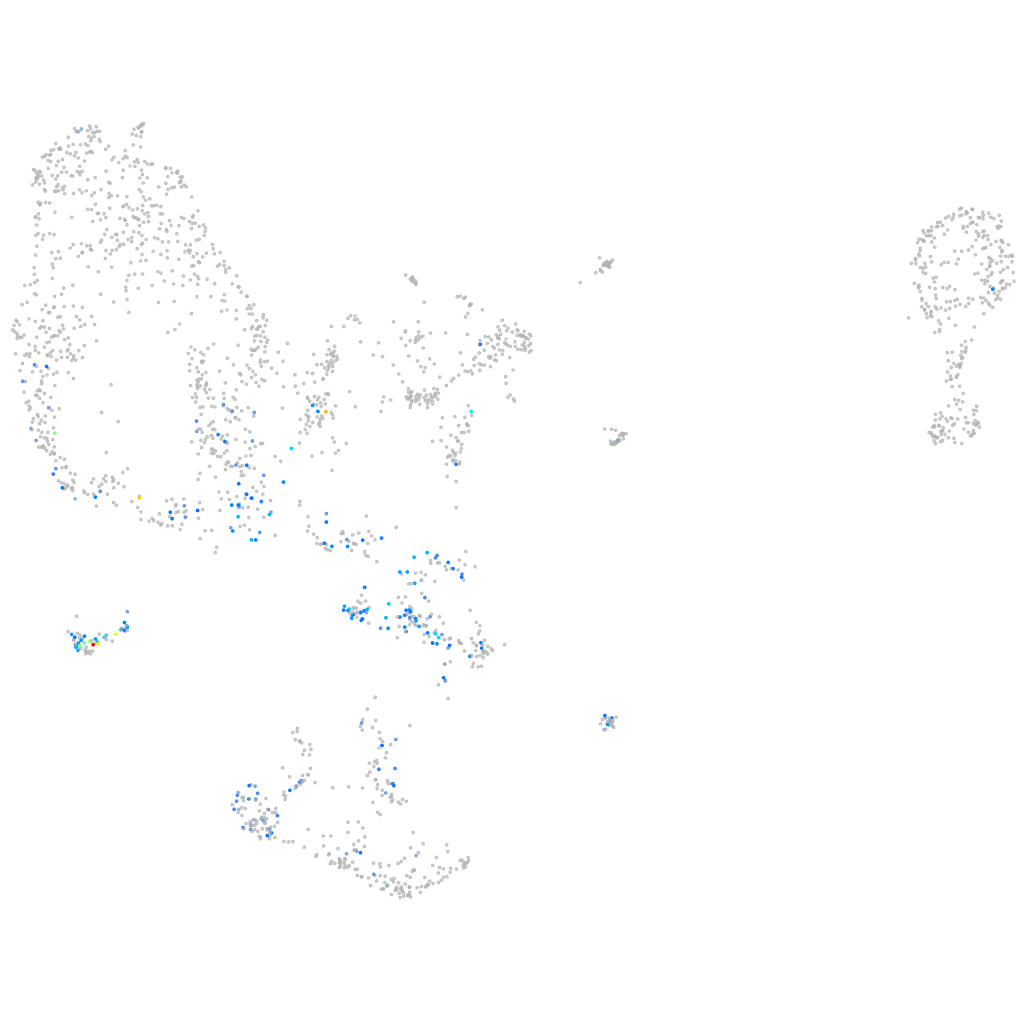
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 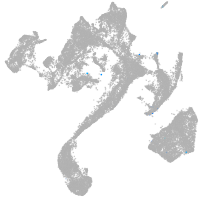 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 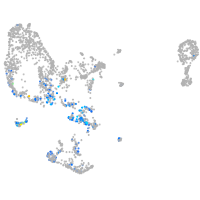 |
neural |
spinal cord / glia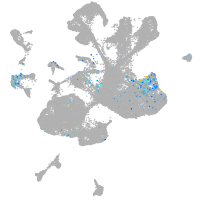 |
eye |
taste / olfactory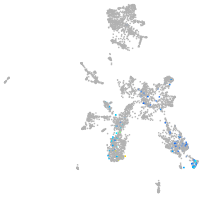 |
otic / lateral line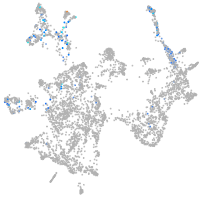 |
epidermis |
periderm |
fin |
ionocytes / mucous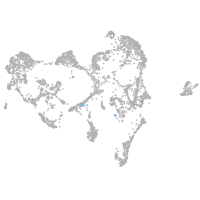 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| spata18 | 0.407 | gcshb | -0.156 |
| tsga10 | 0.394 | gapdh | -0.148 |
| dnali1 | 0.390 | rplp2 | -0.142 |
| dydc2 | 0.381 | slc16a1b | -0.141 |
| spata4 | 0.377 | epdl2 | -0.140 |
| zgc:55461 | 0.375 | agxtb | -0.139 |
| daw1 | 0.374 | alpl | -0.139 |
| si:ch211-163l21.7 | 0.369 | rps6 | -0.137 |
| ribc2 | 0.367 | cx32.3 | -0.135 |
| spag6 | 0.364 | dpys | -0.135 |
| ropn1l | 0.364 | aco1 | -0.134 |
| stpg2 | 0.362 | pnp6 | -0.134 |
| cfap126 | 0.360 | gpt2l | -0.133 |
| cfap20 | 0.359 | phyhd1 | -0.133 |
| cetn4 | 0.356 | slc5a12 | -0.130 |
| cfap45 | 0.354 | gstt1a | -0.129 |
| tekt1 | 0.352 | slc26a6l | -0.129 |
| ttc25 | 0.350 | si:ch211-139a5.9 | -0.129 |
| tpgs2 | 0.350 | slc20a1a | -0.128 |
| rsph9 | 0.349 | cubn | -0.128 |
| capsla | 0.348 | ctsz | -0.127 |
| crocc2 | 0.348 | upb1 | -0.127 |
| cluap1 | 0.348 | asl | -0.127 |
| irak1bp1 | 0.346 | akr7a3 | -0.127 |
| ccdc114 | 0.346 | g6pca.2 | -0.127 |
| ccdc151 | 0.343 | ctsla | -0.126 |
| ccdc173 | 0.341 | rgn | -0.126 |
| tekt4 | 0.340 | slc22a4 | -0.126 |
| ncs1a | 0.339 | slc22a13b | -0.125 |
| sept8b | 0.338 | si:dkey-33i11.9 | -0.125 |
| catip | 0.338 | rpl3 | -0.124 |
| nme5 | 0.337 | ASS1 | -0.124 |
| si:ch1073-416d2.4 | 0.337 | rps18 | -0.124 |
| mycbp | 0.335 | slc34a1a | -0.123 |
| drc3 | 0.334 | hao1 | -0.123 |