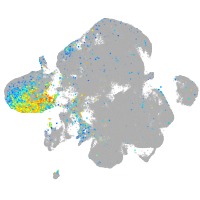
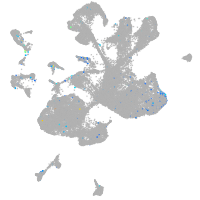
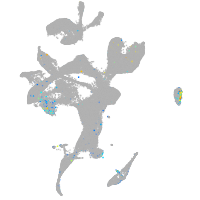
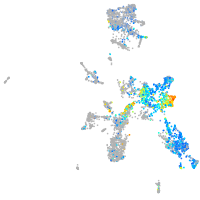
distal-less homeobox 3b
ZFIN 
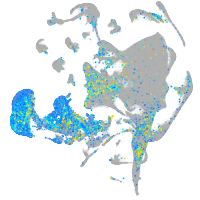
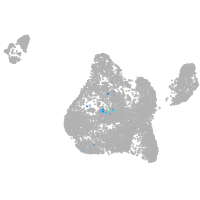
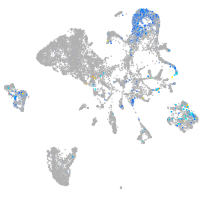
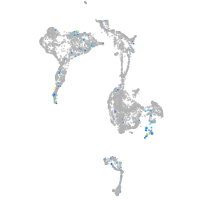
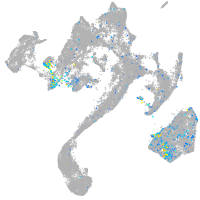
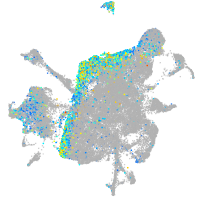
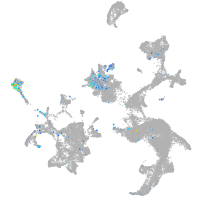
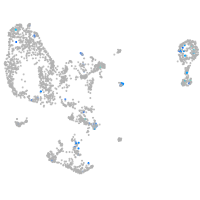
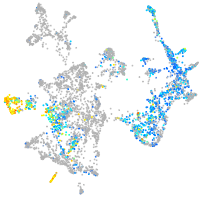
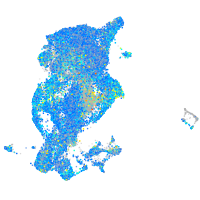
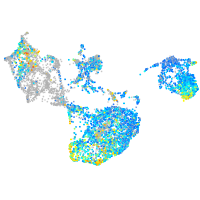
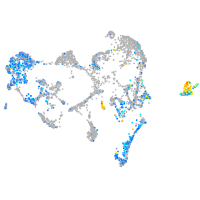
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| foxi1 | 0.575 | rpl37 | -0.354 |
| apoc1 | 0.568 | rps10 | -0.353 |
| apoeb | 0.545 | zgc:114188 | -0.337 |
| ved | 0.509 | h3f3a | -0.323 |
| tp63 | 0.473 | ptmaa | -0.310 |
| si:ch211-152c2.3 | 0.439 | hnrnpa0l | -0.293 |
| tagln2 | 0.437 | fabp3 | -0.283 |
| LOC108190024 | 0.429 | tmsb4x | -0.267 |
| crabp2b | 0.428 | marcksl1a | -0.257 |
| klf17 | 0.425 | rps17 | -0.251 |
| hspb1 | 0.413 | tuba1c | -0.249 |
| bambia | 0.406 | CR383676.1 | -0.233 |
| prdm1a | 0.405 | ppiab | -0.227 |
| vox | 0.403 | si:ch1073-429i10.3.1 | -0.222 |
| cdh1 | 0.396 | nme2b.1 | -0.221 |
| tfap2c | 0.388 | nova2 | -0.211 |
| apela | 0.387 | zgc:158463 | -0.206 |
| nr6a1a | 0.387 | elavl3 | -0.197 |
| NC-002333.4 | 0.382 | gpm6aa | -0.197 |
| bmp7a | 0.374 | rtn1a | -0.197 |
| bmp2b | 0.362 | cct2 | -0.193 |
| klf2a | 0.359 | stmn1b | -0.180 |
| akap12b | 0.348 | mt-nd1 | -0.180 |
| BX927258.1 | 0.347 | h3f3c | -0.179 |
| dnmt3bb.1 | 0.343 | celf2 | -0.172 |
| COX7A2 | 0.335 | COX3 | -0.165 |
| cdx4 | 0.333 | eef1b2 | -0.165 |
| alcamb | 0.329 | tmsb | -0.164 |
| epcam | 0.326 | calm1a | -0.163 |
| fbl | 0.324 | mt-atp6 | -0.163 |
| shisa2a | 0.322 | fam168a | -0.162 |
| stm | 0.320 | prdx2 | -0.161 |
| si:dkey-66i24.9 | 0.316 | ckbb | -0.158 |
| ovol1b | 0.312 | ccni | -0.157 |
| mir205 | 0.308 | wu:fb55g09 | -0.154 |