DLC1 Rho GTPase activating protein
ZFIN 
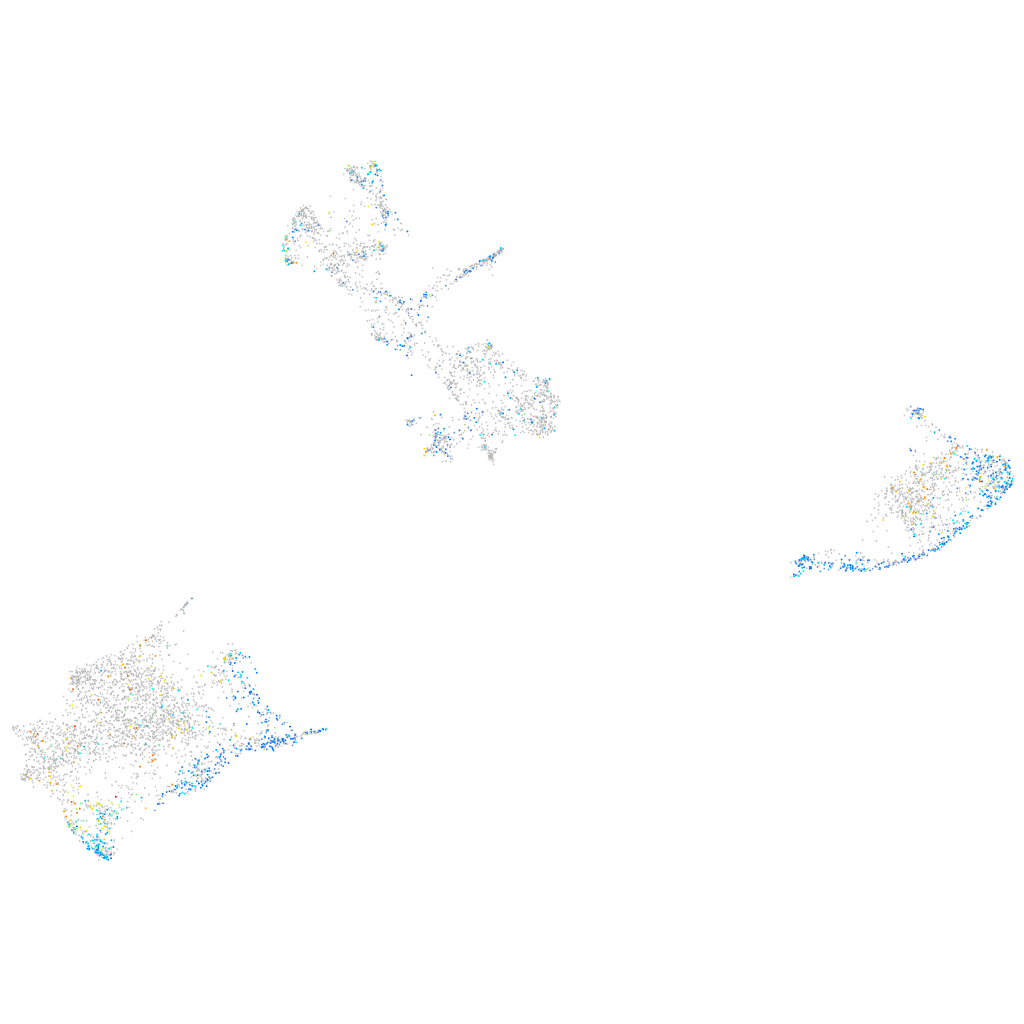
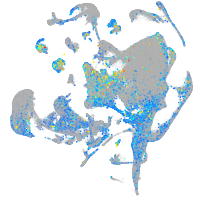
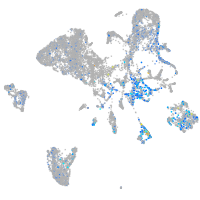
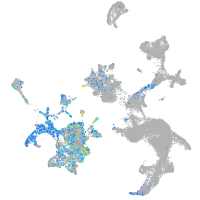
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gadd45bb | 0.190 | ptmaa | -0.106 |
| si:zfos-943e10.1 | 0.184 | tuba1c | -0.097 |
| opn5 | 0.182 | hbbe1.3 | -0.089 |
| trpm1b | 0.182 | mdkb | -0.089 |
| inka1b | 0.180 | si:ch211-222l21.1 | -0.087 |
| SPAG9 | 0.175 | hbae3 | -0.087 |
| socs3a | 0.172 | tmsb | -0.081 |
| pim1 | 0.171 | hbae1.1 | -0.076 |
| sdc4 | 0.169 | gpm6aa | -0.076 |
| tfap2e | 0.168 | elavl3 | -0.076 |
| sik1 | 0.168 | gapdhs | -0.074 |
| klf6a | 0.167 | fabp7a | -0.073 |
| arhgef33 | 0.165 | si:ch73-1a9.3 | -0.073 |
| tspan36 | 0.163 | ptmab | -0.070 |
| id3 | 0.159 | nova2 | -0.069 |
| bace2 | 0.159 | hmgn2 | -0.069 |
| ctsla | 0.159 | CU467822.1 | -0.068 |
| cited4b | 0.156 | hmgb1b | -0.068 |
| crema | 0.155 | stmn1b | -0.068 |
| bhlhe41 | 0.154 | gng3 | -0.067 |
| slc3a2a | 0.153 | epb41a | -0.066 |
| mitfa | 0.152 | fez1 | -0.062 |
| adipor2 | 0.152 | paics | -0.061 |
| ddit3 | 0.151 | stx1b | -0.059 |
| myo5aa | 0.150 | tuba1a | -0.058 |
| ier2a | 0.149 | sncb | -0.058 |
| pcdh10a | 0.149 | defbl1 | -0.057 |
| mlpha | 0.148 | hbbe1.1 | -0.056 |
| si:ch211-195b13.1 | 0.146 | snap25a | -0.056 |
| wsb1 | 0.145 | zc4h2 | -0.055 |
| rasd1 | 0.144 | nptna | -0.055 |
| kita | 0.143 | efnb3b | -0.054 |
| klf15 | 0.143 | uraha | -0.054 |
| CR383676.1 | 0.142 | hbae1.3 | -0.054 |
| cdh1 | 0.141 | foxp4 | -0.053 |