DEAH (Asp-Glu-Ala-His) box polypeptide 37
ZFIN 
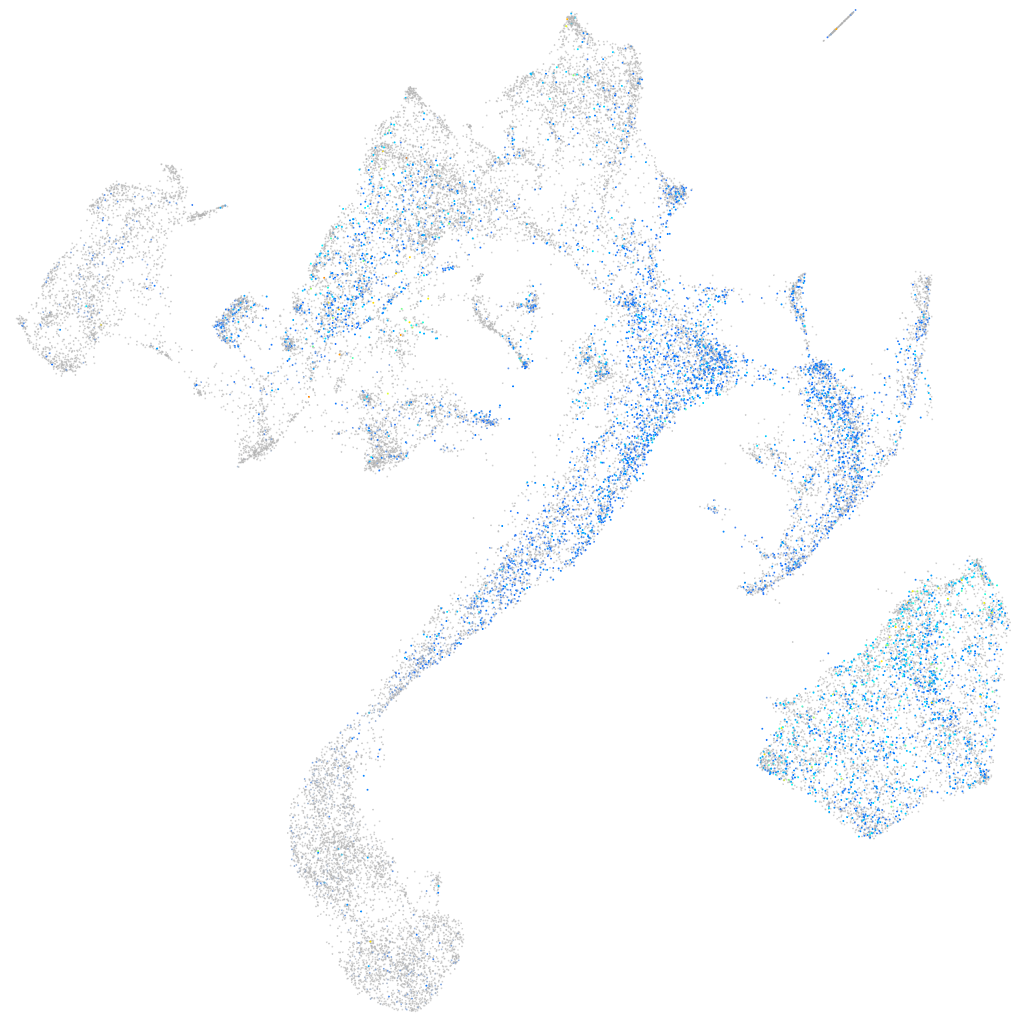
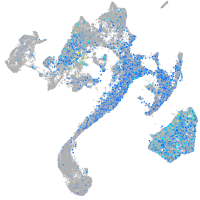
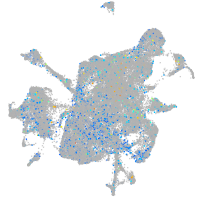
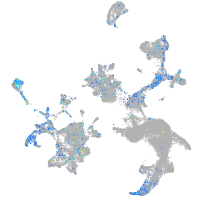
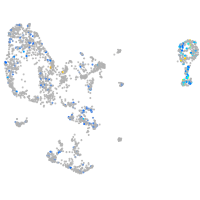
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.291 | actc1b | -0.191 |
| ncl | 0.285 | ckma | -0.189 |
| nop58 | 0.281 | ak1 | -0.185 |
| snu13b | 0.272 | eno3 | -0.184 |
| nop56 | 0.272 | ckmb | -0.184 |
| nop2 | 0.266 | pvalb1 | -0.180 |
| fbl | 0.265 | neb | -0.179 |
| dkc1 | 0.264 | tnnc2 | -0.179 |
| nhp2 | 0.253 | actn3a | -0.177 |
| setb | 0.247 | si:ch73-367p23.2 | -0.176 |
| ebna1bp2 | 0.247 | atp2a1 | -0.174 |
| rrp1 | 0.240 | tpi1b | -0.173 |
| bms1 | 0.239 | bhmt | -0.173 |
| nop10 | 0.238 | smyd1a | -0.173 |
| rsl1d1 | 0.237 | pvalb2 | -0.173 |
| hnrnpabb | 0.236 | ldb3b | -0.173 |
| ssb | 0.235 | tmod4 | -0.171 |
| wdr43 | 0.234 | si:dkey-16p21.8 | -0.171 |
| si:ch211-217k17.7 | 0.233 | nme2b.2 | -0.170 |
| ptges3b | 0.233 | si:ch211-266g18.10 | -0.169 |
| rpl7l1 | 0.232 | gapdh | -0.169 |
| syncrip | 0.232 | myom1a | -0.168 |
| gnl3 | 0.232 | mylpfa | -0.168 |
| mybbp1a | 0.231 | actn3b | -0.168 |
| eif5a2 | 0.230 | pgam2 | -0.167 |
| snrpb | 0.230 | aldoab | -0.167 |
| pprc1 | 0.228 | tnnt3a | -0.164 |
| ppan | 0.225 | eno1a | -0.164 |
| lyar | 0.224 | tnnt3b | -0.163 |
| pes | 0.224 | acta1b | -0.163 |
| ddx27 | 0.223 | mylz3 | -0.163 |
| mak16 | 0.223 | casq2 | -0.162 |
| hmga1a | 0.222 | prx | -0.162 |
| khdrbs1a | 0.222 | pmp22b | -0.161 |
| cbx3a | 0.222 | ank1a | -0.161 |