desert hedgehog signaling molecule
ZFIN 
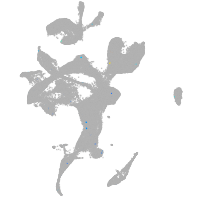
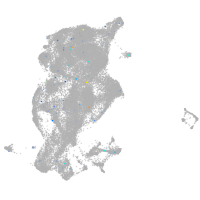
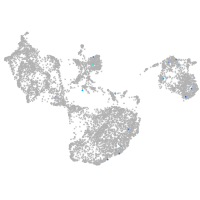
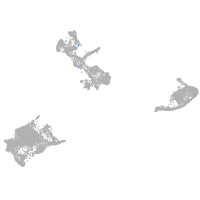
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101885599 | 0.321 | gapdh | -0.078 |
| slc26a10 | 0.290 | ahcy | -0.072 |
| LOC110439394 | 0.249 | gatm | -0.071 |
| CR932999.1 | 0.247 | gamt | -0.069 |
| sim1b | 0.242 | bhmt | -0.066 |
| anxa3b | 0.241 | fbp1b | -0.064 |
| myofl | 0.240 | hspe1 | -0.062 |
| spock3 | 0.238 | apoc2 | -0.061 |
| abca12 | 0.238 | aqp12 | -0.061 |
| si:ch211-274p24.4 | 0.234 | apoa4b.1 | -0.060 |
| RASA2 | 0.233 | mat1a | -0.059 |
| ceacam1 | 0.228 | dap | -0.059 |
| cabp2b | 0.225 | afp4 | -0.058 |
| CABZ01055365.1 | 0.215 | si:dkey-16p21.8 | -0.058 |
| sftpba | 0.214 | mdh1aa | -0.058 |
| FAM163B (1 of many) | 0.213 | scp2a | -0.057 |
| RF00322 | 0.212 | apoa1b | -0.056 |
| actn1 | 0.211 | suclg1 | -0.055 |
| znf644b | 0.209 | atp5mc3b | -0.055 |
| lama5 | 0.208 | grhprb | -0.054 |
| pmp22b | 0.207 | sult2st2 | -0.054 |
| ahnak | 0.203 | apoc1 | -0.053 |
| palm2 | 0.202 | gcshb | -0.053 |
| BX927336.1 | 0.199 | gpx4a | -0.052 |
| capn2a | 0.197 | gstt1a | -0.052 |
| FP102018.1 | 0.195 | suclg2 | -0.052 |
| glis2a | 0.193 | agxtb | -0.051 |
| cygb1 | 0.190 | slc27a2a | -0.051 |
| LOC101885772 | 0.189 | apobb.1 | -0.051 |
| mxra8b | 0.187 | gpd1b | -0.051 |
| scamp2l | 0.185 | pnp4b | -0.050 |
| LOC101885062 | 0.184 | agxta | -0.050 |
| gna15.1 | 0.184 | g6pca.2 | -0.050 |
| crygm2d10 | 0.182 | abat | -0.050 |
| spaca4l | 0.179 | fabp3 | -0.049 |