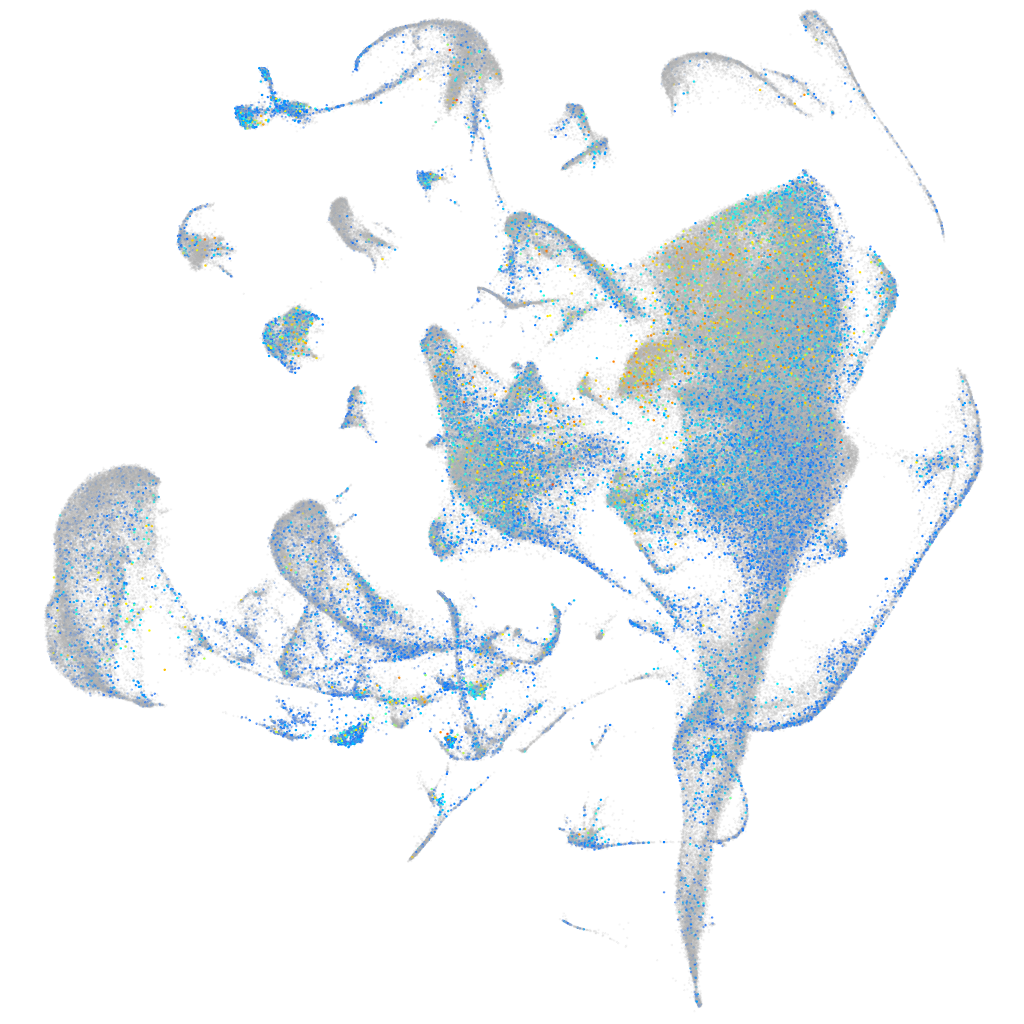Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| laptm5 | 0.055 | aldob | -0.034 |
| coro1a | 0.053 | si:dkey-16p21.8 | -0.028 |
| marcksl1a | 0.053 | cyt1l | -0.027 |
| ceacam1 | 0.051 | wu:fa03e10 | -0.027 |
| itgb2 | 0.051 | krtt1c19e | -0.026 |
| rgs13 | 0.051 | aep1 | -0.025 |
| fcer1gl | 0.050 | hrc | -0.025 |
| ccr9a | 0.049 | krt5 | -0.025 |
| foxi3b | 0.049 | eevs | -0.024 |
| si:ch211-102c2.4 | 0.049 | fut9d | -0.024 |
| spi1b | 0.049 | tcnbb | -0.024 |
| klf6a | 0.048 | casq2 | -0.023 |
| cldnh | 0.047 | icn2 | -0.023 |
| LOC100536213 | 0.047 | krt17 | -0.023 |
| ptprc | 0.047 | lye | -0.023 |
| sat1a.2 | 0.047 | zgc:193505 | -0.023 |
| si:dkey-33i11.4 | 0.047 | aldoab | -0.022 |
| FERMT3 (1 of many) | 0.047 | cav3 | -0.022 |
| gmfg | 0.047 | ckmb | -0.022 |
| zgc:64051 | 0.047 | cyt1 | -0.022 |
| grap2b | 0.046 | desma | -0.022 |
| itgae.2 | 0.046 | gapdh | -0.022 |
| lcp1 | 0.046 | gpd1b | -0.022 |
| rhogb | 0.046 | si:ch211-266g18.10 | -0.022 |
| adgrg3 | 0.045 | si:ch73-52f15.5 | -0.022 |
| cd81a | 0.045 | srl | -0.022 |
| glipr1a | 0.045 | ugp2a | -0.022 |
| gpr183a | 0.045 | atp2a1 | -0.022 |
| il1b | 0.045 | actn3a | -0.021 |
| mpeg1.1 | 0.045 | actn3b | -0.021 |
| ptpn6 | 0.045 | agr1 | -0.021 |
| fcer1g | 0.045 | anxa1c | -0.021 |
| anxa3b | 0.044 | ckma | -0.021 |
| cmklr1 | 0.044 | ckmt2b | -0.021 |
| CU499330.1 | 0.044 | grhprb | -0.021 |