dihydrofolate reductase
ZFIN 
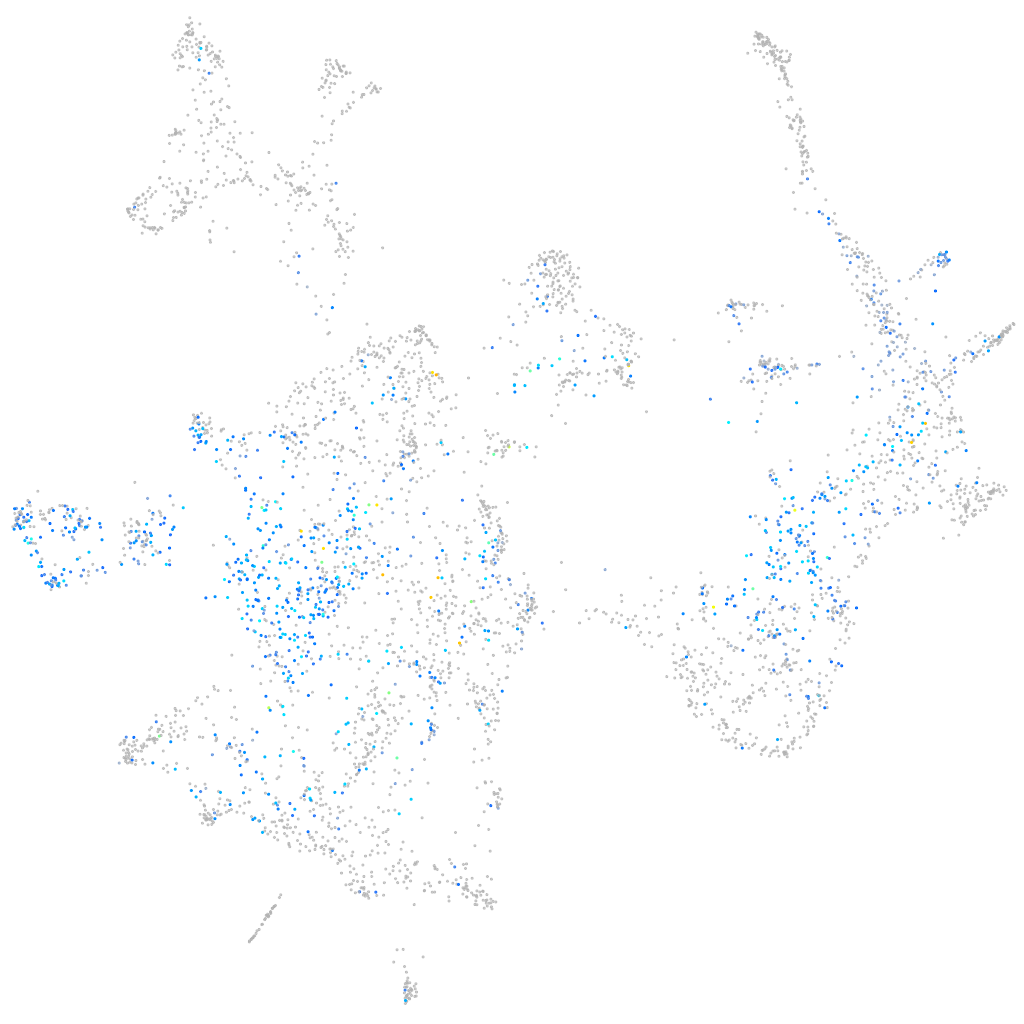
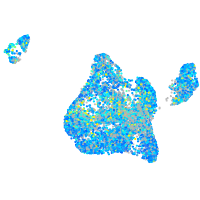
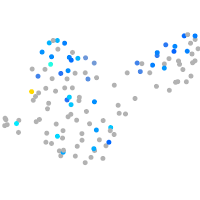
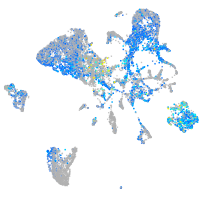
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.615 | atp2b1a | -0.187 |
| pcna | 0.614 | h1f0 | -0.186 |
| rpa2 | 0.581 | tob1b | -0.185 |
| stmn1a | 0.555 | calm1a | -0.183 |
| zgc:110540 | 0.553 | nupr1a | -0.179 |
| rrm2 | 0.548 | otofb | -0.173 |
| rrm1 | 0.544 | tmem59 | -0.170 |
| chaf1a | 0.533 | nptna | -0.169 |
| nasp | 0.516 | atp1a3b | -0.167 |
| fen1 | 0.515 | gpx2 | -0.166 |
| rpa3 | 0.514 | calml4a | -0.165 |
| CABZ01005379.1 | 0.507 | rnasekb | -0.164 |
| cks1b | 0.499 | kif1aa | -0.164 |
| slbp | 0.498 | cd164l2 | -0.163 |
| mibp | 0.489 | dnajc5b | -0.162 |
| orc4 | 0.486 | osbpl1a | -0.160 |
| si:ch211-156b7.4 | 0.486 | CR383676.1 | -0.159 |
| lig1 | 0.474 | ip6k2a | -0.159 |
| si:dkey-6i22.5 | 0.471 | CR925719.1 | -0.158 |
| prim2 | 0.471 | lrrc73 | -0.158 |
| dnajc9 | 0.466 | eno1a | -0.158 |
| tubb2b | 0.461 | tspan13b | -0.157 |
| dek | 0.459 | atp6v1e1b | -0.156 |
| rnaseh2a | 0.458 | pcsk5a | -0.155 |
| banf1 | 0.457 | si:ch73-15n15.3 | -0.155 |
| cdca5 | 0.455 | atp1b2b | -0.155 |
| asf1ba | 0.452 | COX3 | -0.154 |
| shmt1 | 0.450 | emb | -0.154 |
| unga | 0.448 | apba1a | -0.153 |
| trip13 | 0.448 | ckbb | -0.152 |
| anp32b | 0.447 | slc17a8 | -0.152 |
| esco2 | 0.447 | myo6b | -0.151 |
| zgc:110216 | 0.445 | gipc3 | -0.151 |
| mcm7 | 0.444 | aldocb | -0.150 |
| rfc4 | 0.440 | rbm24a | -0.150 |