DEK proto-oncogene
ZFIN 
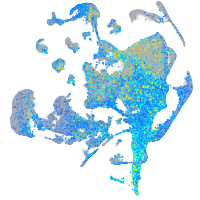
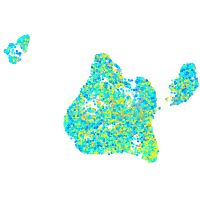
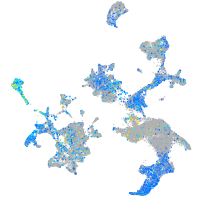
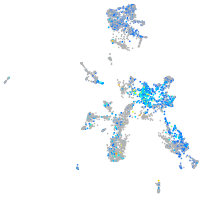
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrm1 | 0.554 | tob1b | -0.166 |
| dut | 0.536 | COX3 | -0.162 |
| pcna | 0.531 | h1f0 | -0.156 |
| cks1b | 0.515 | atp1a3b | -0.143 |
| rrm2 | 0.509 | tmem59 | -0.142 |
| chaf1a | 0.506 | otofb | -0.141 |
| stmn1a | 0.504 | nptna | -0.140 |
| ccna2 | 0.495 | cd164l2 | -0.139 |
| cdca5 | 0.495 | nupr1a | -0.138 |
| zgc:110216 | 0.495 | rnasekb | -0.137 |
| mibp | 0.494 | gpx2 | -0.136 |
| zgc:110540 | 0.491 | pvalb8 | -0.136 |
| mki67 | 0.489 | eno1a | -0.133 |
| rpa2 | 0.486 | pvalb2 | -0.132 |
| ncapg | 0.482 | CR383676.1 | -0.132 |
| zgc:165555.3 | 0.481 | calml4a | -0.131 |
| aurkb | 0.477 | si:dkey-16p21.8 | -0.131 |
| CABZ01005379.1 | 0.465 | emb | -0.130 |
| si:dkey-6i22.5 | 0.464 | ube2h | -0.130 |
| tubb2b | 0.463 | osbpl1a | -0.130 |
| anp32b | 0.463 | CR925719.1 | -0.130 |
| dhfr | 0.459 | gabarapl2 | -0.129 |
| hmgb2a | 0.459 | gapdhs | -0.128 |
| fbxo5 | 0.457 | calm1a | -0.128 |
| mad2l1 | 0.455 | kif1aa | -0.128 |
| cdk1 | 0.455 | atp6v1e1b | -0.127 |
| esco2 | 0.446 | b2ml | -0.127 |
| smc2 | 0.445 | dnajc5b | -0.127 |
| banf1 | 0.445 | apba1a | -0.126 |
| zgc:110425 | 0.444 | pvalb1 | -0.125 |
| prim2 | 0.441 | ckbb | -0.125 |
| lbr | 0.434 | si:ch73-15n15.3 | -0.124 |
| nasp | 0.433 | lrrc73 | -0.123 |
| h3f3a | 0.430 | atp2b1a | -0.123 |
| rpa3 | 0.428 | vamp2 | -0.122 |