DEAD (Asp-Glu-Ala-Asp) box helicase 6
ZFIN 
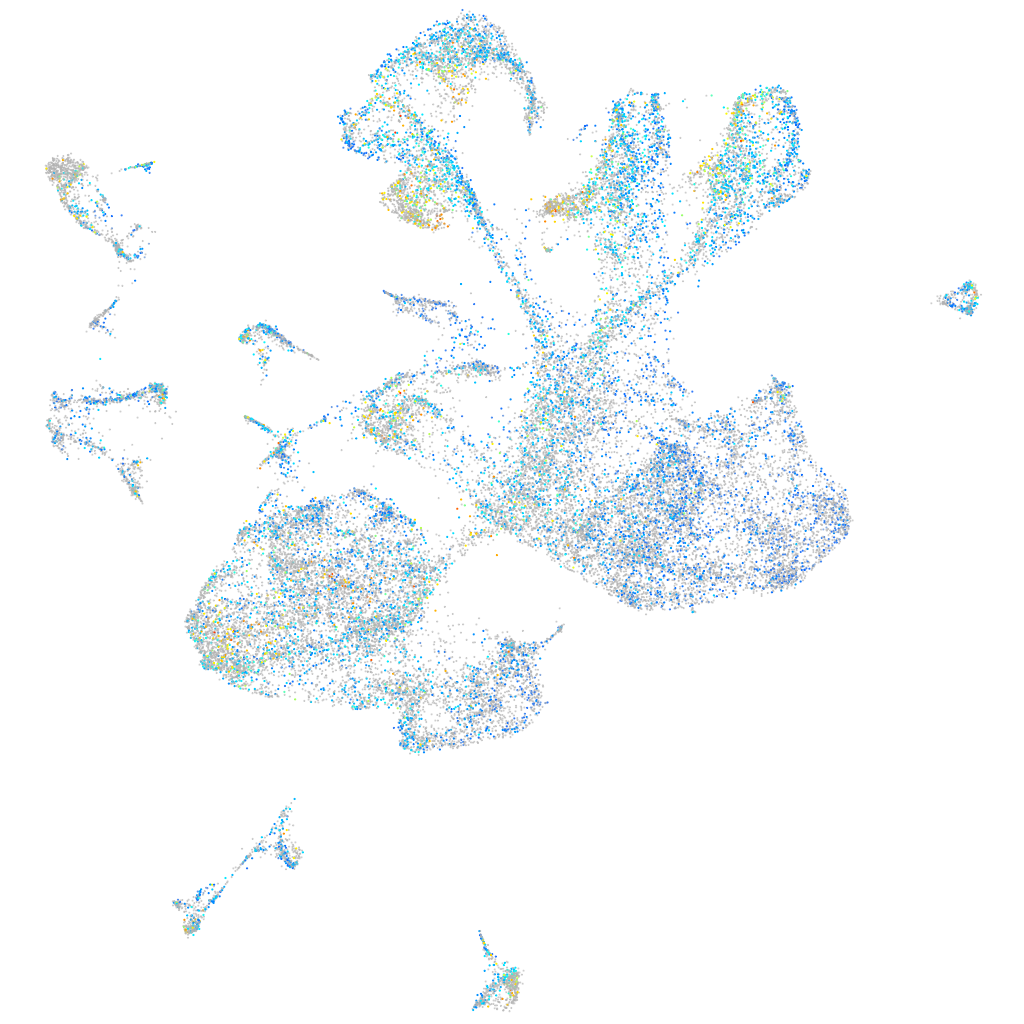
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl4 | 0.175 | XLOC-003690 | -0.142 |
| stx1b | 0.169 | hmgb2a | -0.134 |
| gng3 | 0.165 | her15.1 | -0.127 |
| cplx2 | 0.164 | XLOC-003689 | -0.124 |
| snap25a | 0.163 | cldn5a | -0.117 |
| vamp2 | 0.163 | id1 | -0.116 |
| stxbp1a | 0.163 | pcna | -0.110 |
| sncb | 0.161 | zgc:56493 | -0.109 |
| atp6v0cb | 0.159 | her2 | -0.109 |
| ywhag2 | 0.155 | rplp1 | -0.109 |
| stmn2a | 0.155 | dut | -0.107 |
| sncgb | 0.154 | si:dkey-151g10.6 | -0.107 |
| rtn1b | 0.148 | her12 | -0.106 |
| ywhah | 0.147 | rps20 | -0.104 |
| elavl3 | 0.146 | ran | -0.104 |
| zgc:65894 | 0.145 | rpl23 | -0.103 |
| atp1a3a | 0.145 | XLOC-003692 | -0.102 |
| syt2a | 0.144 | rps7 | -0.101 |
| stmn1b | 0.143 | selenoh | -0.101 |
| rbfox1 | 0.141 | rpl10a | -0.100 |
| myt1b | 0.141 | sox3 | -0.100 |
| calm1a | 0.140 | rps2 | -0.100 |
| eno2 | 0.140 | stmn1a | -0.100 |
| tuba2 | 0.136 | rpl26 | -0.099 |
| map1aa | 0.136 | rplp0 | -0.099 |
| nsg2 | 0.136 | rps15a | -0.099 |
| tmem59l | 0.136 | sdc4 | -0.098 |
| ndrg4 | 0.135 | eef1a1l1 | -0.098 |
| cdk5r1b | 0.135 | chaf1a | -0.097 |
| cplx2l | 0.135 | COX7A2 (1 of many) | -0.097 |
| atp1b2a | 0.134 | rpl11 | -0.097 |
| atp6v1e1b | 0.133 | mdka | -0.096 |
| sypb | 0.133 | rpl7a | -0.096 |
| dpysl3 | 0.133 | rpl12 | -0.096 |
| gng2 | 0.132 | ranbp1 | -0.094 |