DEAD (Asp-Glu-Ala-Asp) box polypeptide 43
ZFIN 
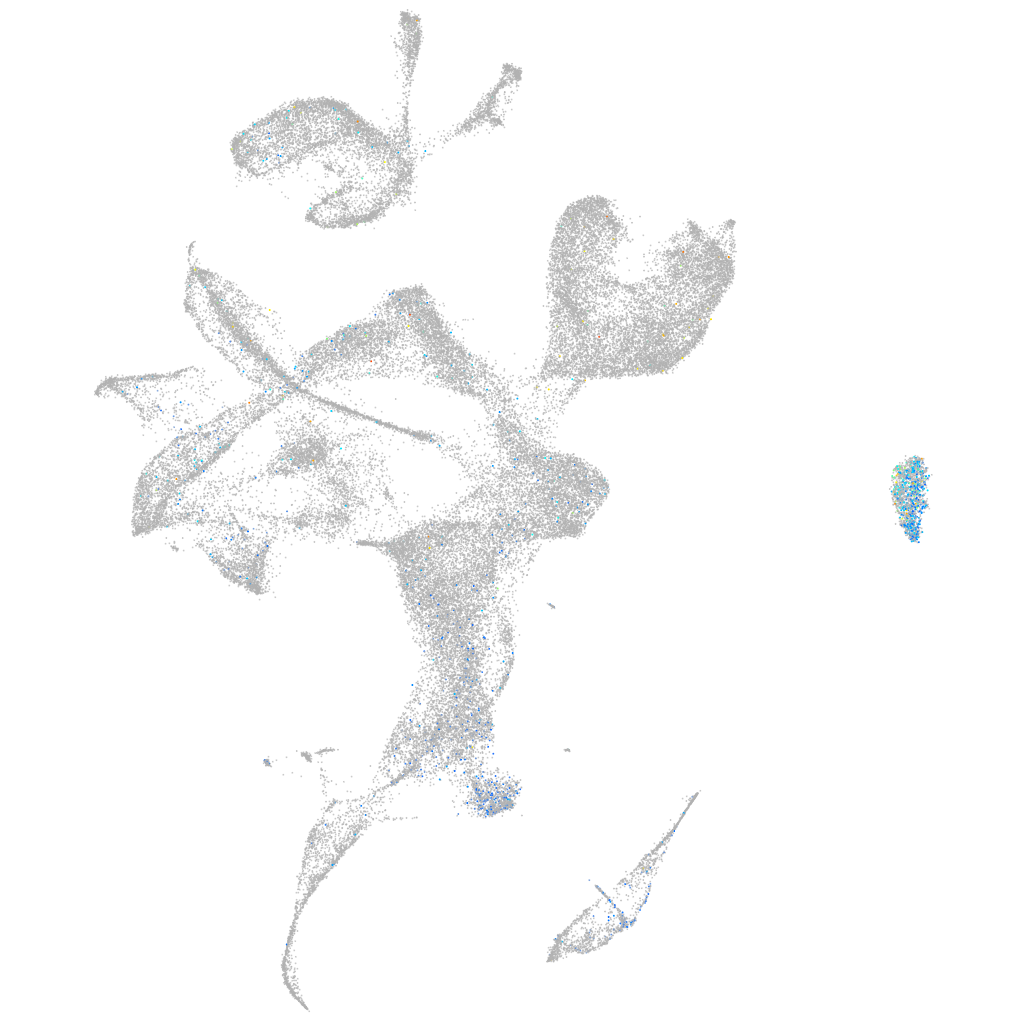
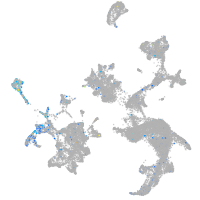
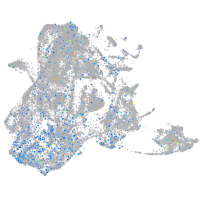
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.259 | rpl37 | -0.119 |
| shisa2a | 0.235 | rps10 | -0.115 |
| si:ch211-152c2.3 | 0.234 | CR383676.1 | -0.110 |
| stm | 0.226 | ptmaa | -0.107 |
| apoeb | 0.219 | zgc:114188 | -0.103 |
| cyp26a1 | 0.218 | h3f3a | -0.101 |
| tdgf1 | 0.210 | ppiab | -0.094 |
| hesx1 | 0.207 | hnrnpa0l | -0.086 |
| nr6a1a | 0.205 | COX3 | -0.085 |
| sox19a | 0.202 | rps17 | -0.079 |
| aldob | 0.197 | zgc:158463 | -0.079 |
| LOC108190024 | 0.194 | gpm6aa | -0.078 |
| zmp:0000000624 | 0.192 | marcksl1a | -0.077 |
| s100a1 | 0.192 | si:ch1073-429i10.3.1 | -0.074 |
| COX7A2 | 0.191 | tuba1c | -0.074 |
| asb11 | 0.182 | mt-atp6 | -0.073 |
| pkdccb | 0.182 | ckbb | -0.072 |
| polr3gla | 0.181 | mt-nd1 | -0.068 |
| foxd5 | 0.180 | gpm6ab | -0.068 |
| pprc1 | 0.178 | pvalb2 | -0.067 |
| NC-002333.4 | 0.176 | mt-co2 | -0.066 |
| mybbp1a | 0.174 | actc1b | -0.066 |
| gnl3 | 0.174 | pvalb1 | -0.065 |
| bms1 | 0.173 | h3f3c | -0.064 |
| npm1a | 0.172 | cadm3 | -0.063 |
| alcamb | 0.171 | fabp3 | -0.059 |
| chrd | 0.168 | zgc:153409 | -0.059 |
| dkc1 | 0.166 | fabp7a | -0.058 |
| nop2 | 0.165 | hbbe1.3 | -0.057 |
| fbl | 0.165 | gapdhs | -0.055 |
| apoc1 | 0.163 | ppdpfb | -0.053 |
| nop58 | 0.163 | rnasekb | -0.051 |
| gar1 | 0.162 | epb41a | -0.051 |
| apela | 0.162 | krt4 | -0.049 |
| sfrp1a | 0.161 | cct2 | -0.049 |