DEAD (Asp-Glu-Ala-Asp) box polypeptide 43
ZFIN 
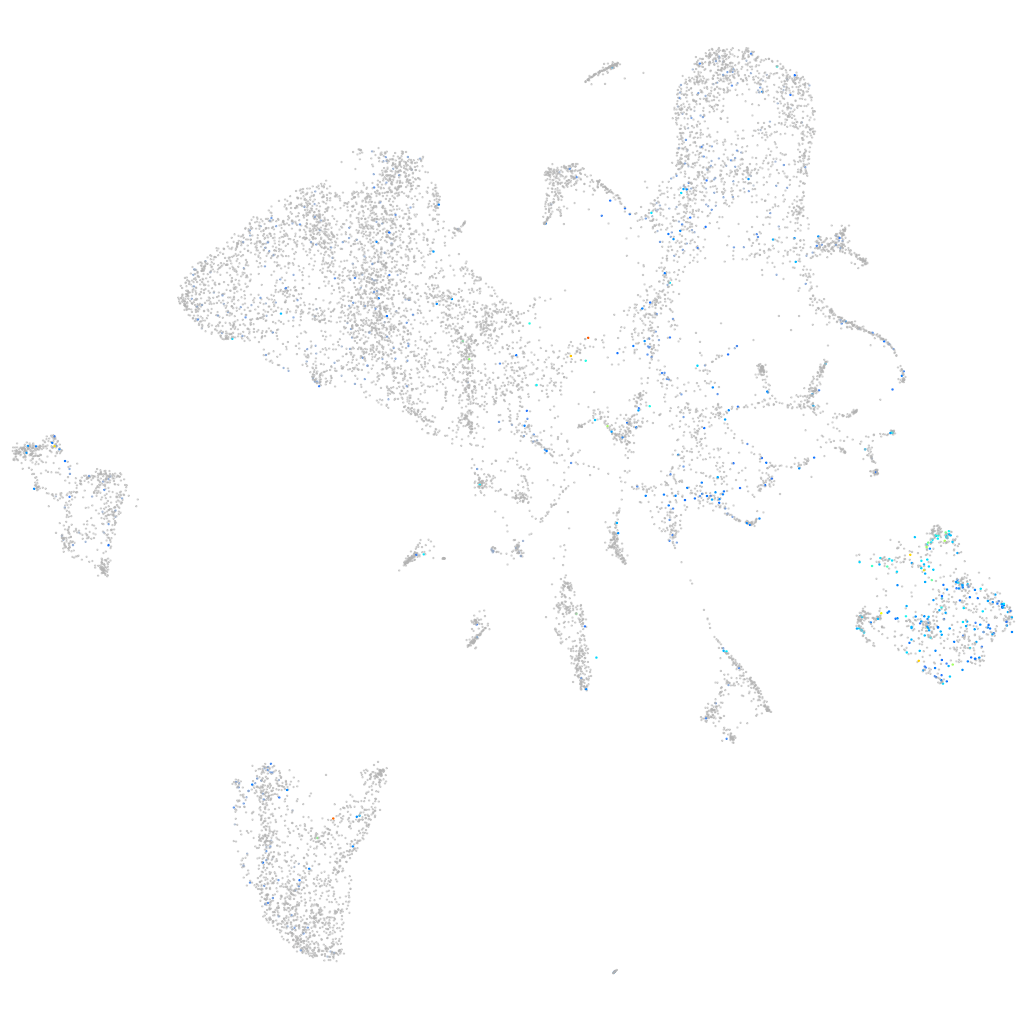
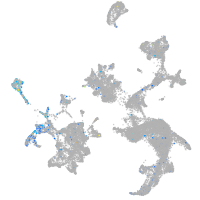
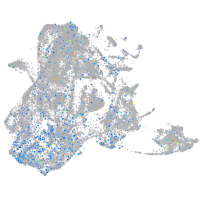
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.262 | rpl37 | -0.218 |
| marcksb | 0.257 | rps10 | -0.212 |
| hnrnpa1b | 0.249 | zgc:114188 | -0.195 |
| si:ch211-152c2.3 | 0.242 | rps17 | -0.175 |
| akap12b | 0.241 | nme2b.1 | -0.162 |
| nucks1a | 0.237 | zgc:92744 | -0.137 |
| cx43.4 | 0.236 | ahcy | -0.136 |
| snrnp70 | 0.235 | gamt | -0.134 |
| hnrnpub | 0.235 | gapdh | -0.130 |
| ppm1g | 0.232 | sod1 | -0.122 |
| hnrnpa1a | 0.231 | eef1da | -0.122 |
| hnrnpm | 0.230 | eno3 | -0.121 |
| anp32e | 0.229 | gatm | -0.118 |
| prpf40a | 0.227 | bhmt | -0.114 |
| si:dkey-66i24.9 | 0.227 | fabp3 | -0.113 |
| vox | 0.225 | prdx2 | -0.112 |
| ncl | 0.224 | apoa1b | -0.111 |
| seta | 0.223 | nupr1b | -0.110 |
| NC-002333.4 | 0.223 | gpx4a | -0.110 |
| nucks1b | 0.223 | atp5if1b | -0.110 |
| ilf3b | 0.223 | tpi1b | -0.109 |
| COX7A2 | 0.223 | suclg1 | -0.109 |
| snrpd3l | 0.223 | pklr | -0.108 |
| acin1a | 0.222 | zgc:158463 | -0.108 |
| sae1 | 0.222 | eef1b2 | -0.107 |
| ppig | 0.222 | dap | -0.107 |
| sp5l | 0.222 | atp5f1b | -0.107 |
| brd3a | 0.221 | COX7A2 (1 of many) | -0.107 |
| smarca4a | 0.219 | apoa2 | -0.106 |
| syncrip | 0.217 | mt-atp6 | -0.105 |
| srrm2 | 0.217 | atp5l | -0.104 |
| anp32a | 0.216 | rplp0 | -0.104 |
| top1l | 0.216 | mdh1aa | -0.104 |
| fbl | 0.216 | mt-nd1 | -0.103 |
| asph | 0.216 | apoa4b.1 | -0.103 |