defective in cullin neddylation 1 domain containing 3
ZFIN 
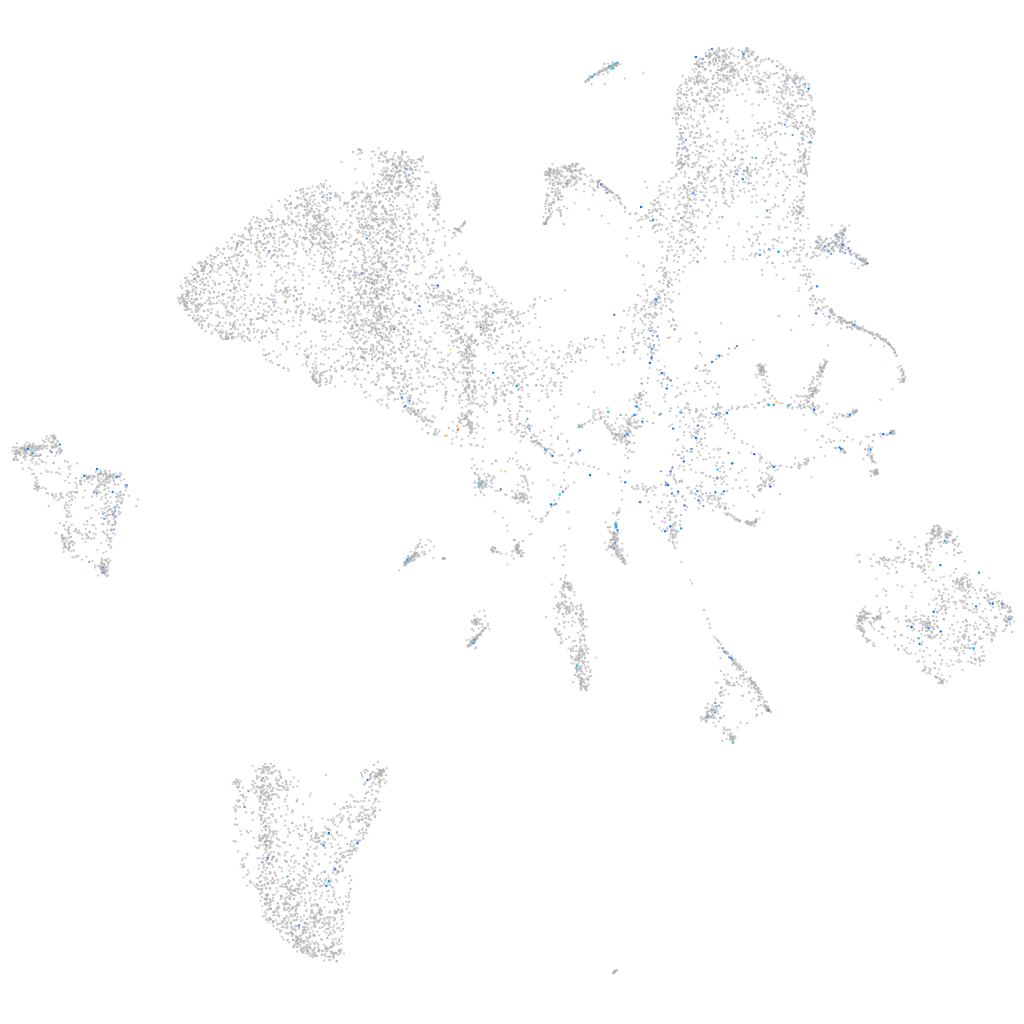
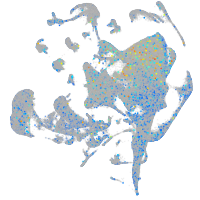
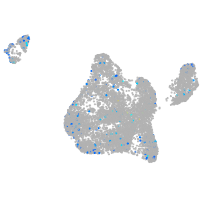
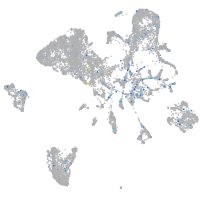
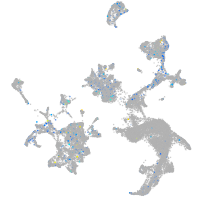
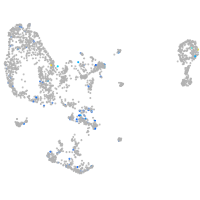
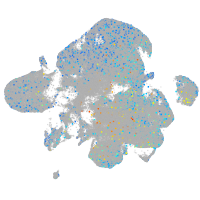
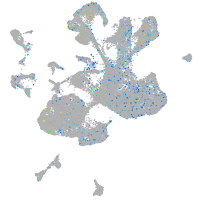
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slitrk5a | 0.180 | gatm | -0.078 |
| urgcp | 0.172 | gamt | -0.078 |
| mir199-2 | 0.162 | bhmt | -0.074 |
| tbkbp1 | 0.161 | mat1a | -0.069 |
| BX927216.1 | 0.148 | agxtb | -0.067 |
| tectb | 0.145 | gapdh | -0.065 |
| gata1a | 0.133 | apoa1b | -0.063 |
| PRMT8 | 0.127 | apoc2 | -0.063 |
| hmx1 | 0.126 | ces2 | -0.062 |
| cx28.6 | 0.121 | apoa2 | -0.062 |
| si:ch211-271g18.4 | 0.116 | afp4 | -0.062 |
| XLOC-025327 | 0.115 | ambp | -0.061 |
| si:cabz01069013.4 | 0.114 | apoc1 | -0.061 |
| igfbp5a | 0.114 | apom | -0.061 |
| zgc:171558 | 0.114 | nupr1b | -0.060 |
| pde6a | 0.113 | fetub | -0.060 |
| LOC101882480 | 0.113 | apoa4b.1 | -0.060 |
| cadpsb | 0.110 | LOC110437731 | -0.060 |
| si:dkeyp-80c12.4 | 0.109 | serpina1 | -0.060 |
| LOC100334340 | 0.109 | tfa | -0.060 |
| BX321877.1 | 0.108 | zgc:123103 | -0.060 |
| BX470259.1 | 0.107 | f2 | -0.059 |
| XLOC-043124 | 0.105 | grhprb | -0.058 |
| TENM2 | 0.104 | fbp1b | -0.058 |
| si:ch211-246e12.3 | 0.102 | gc | -0.058 |
| zgc:109965 | 0.102 | igfbp2a | -0.058 |
| CABZ01084942.1 | 0.101 | c9 | -0.058 |
| XLOC-037017 | 0.100 | aqp12 | -0.058 |
| scdb | 0.098 | pnp4b | -0.057 |
| agap2 | 0.098 | rbp2b | -0.057 |
| CR407590.2 | 0.098 | slc27a2a | -0.057 |
| gabra4 | 0.097 | serpina1l | -0.057 |
| LOC100333313 | 0.093 | hpda | -0.056 |
| LOC101882784 | 0.093 | ttr | -0.056 |
| pax5 | 0.092 | hao1 | -0.056 |