doublecortin-like kinase 2b
ZFIN 
Other cell groups


all cells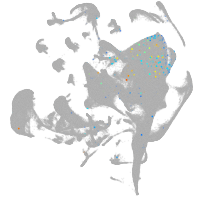 |
blastomeres |
PGCs |
endoderm |
axial mesoderm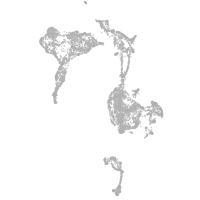 |
muscle  |
mural cells / non-skeletal muscle 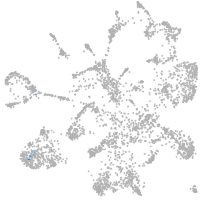 |
mesenchyme 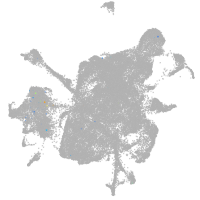 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01081908.1 | 0.125 | hmgb2a | -0.039 |
| gpr6 | 0.124 | anp32e | -0.025 |
| eva1c | 0.090 | pcna | -0.024 |
| alcama | 0.082 | mcm7 | -0.024 |
| elavl3 | 0.077 | msna | -0.023 |
| XLOC-007663 | 0.075 | id1 | -0.022 |
| rbpms2b | 0.074 | nutf2l | -0.022 |
| cxcr4b | 0.072 | ccnd1 | -0.022 |
| XLOC-042485 | 0.070 | CABZ01005379.1 | -0.022 |
| LOC108191789 | 0.070 | stmn1a | -0.021 |
| mllt11 | 0.070 | banf1 | -0.021 |
| LOC110439965 | 0.069 | histh1l | -0.021 |
| XLOC-004872 | 0.069 | lbr | -0.021 |
| tubb5 | 0.068 | lig1 | -0.021 |
| zgc:101715 | 0.066 | fen1 | -0.021 |
| taar20q | 0.066 | rrm1 | -0.020 |
| dpysl3 | 0.065 | rrm2 | -0.020 |
| CR846087.3 | 0.064 | psat1 | -0.020 |
| tmsb | 0.064 | si:ch211-156b7.4 | -0.020 |
| adam22 | 0.063 | msi1 | -0.020 |
| zgc:158291 | 0.061 | mki67 | -0.020 |
| rbfox2 | 0.061 | ccna2 | -0.020 |
| adcyap1b | 0.061 | mcm2 | -0.019 |
| gap43 | 0.061 | ssrp1a | -0.019 |
| si:dkey-280e21.3 | 0.060 | chaf1a | -0.019 |
| LOC110439366 | 0.059 | si:ch73-281n10.2 | -0.019 |
| si:dkeyp-75b4.10 | 0.058 | nop56 | -0.019 |
| klf7b | 0.058 | neurod4 | -0.019 |
| dicp3.3 | 0.058 | cdca7a | -0.018 |
| pbx1a | 0.057 | shmt1 | -0.018 |
| SH3RF3 | 0.057 | ptbp1a | -0.018 |
| zgc:136683 | 0.056 | adi1 | -0.018 |
| nrn1a | 0.056 | jam3b | -0.018 |
| pak1 | 0.056 | si:ch211-214j24.10 | -0.018 |
| mir125b-3 | 0.055 | mibp | -0.018 |




