doublecortin-like kinase 2a
ZFIN 
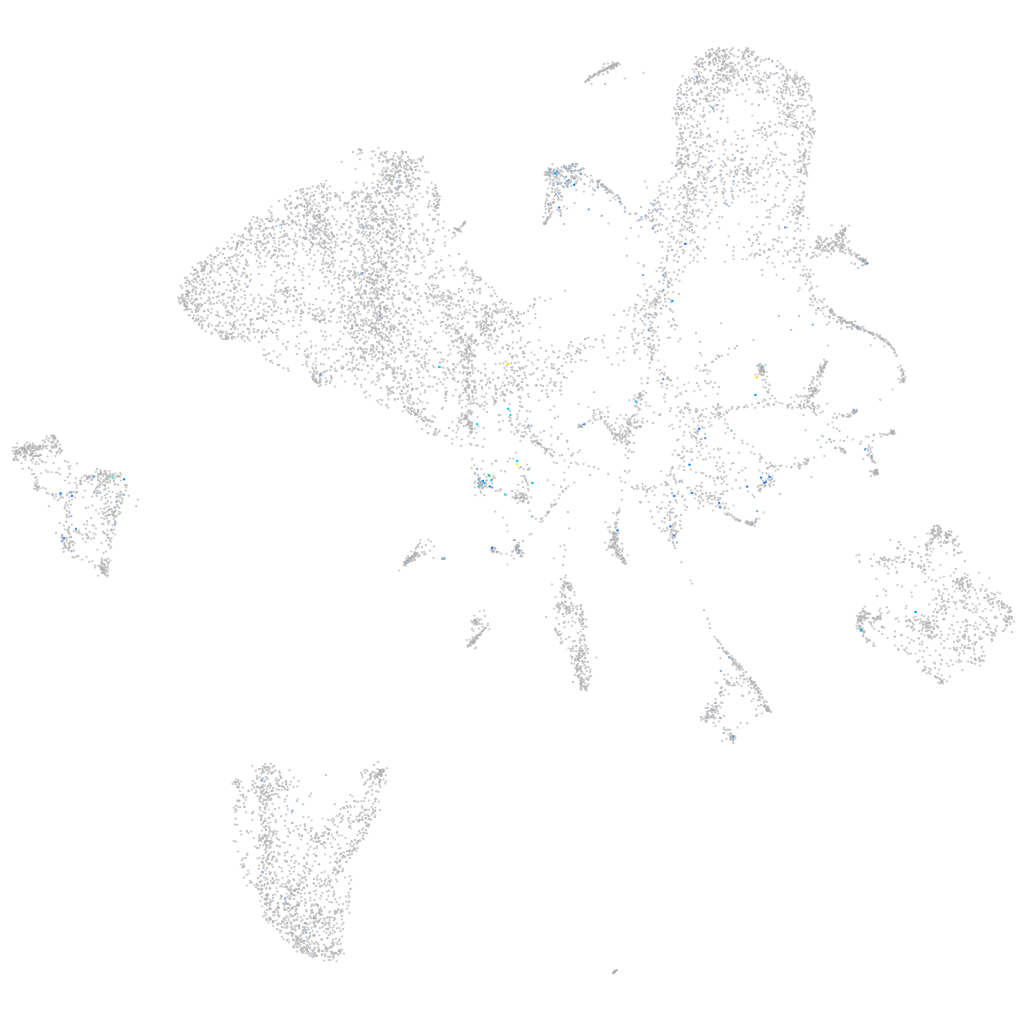
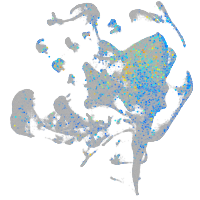
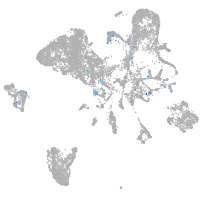
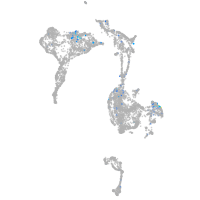
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-043762 | 0.223 | gapdh | -0.078 |
| hes2.1 | 0.223 | gamt | -0.074 |
| tmie | 0.205 | gatm | -0.073 |
| CABZ01087091.1 | 0.205 | aldob | -0.073 |
| XLOC-004644 | 0.191 | nupr1b | -0.070 |
| LOC103909610 | 0.191 | mat1a | -0.070 |
| si:dkey-4c15.16 | 0.183 | fbp1b | -0.066 |
| ptx4 | 0.174 | glud1b | -0.066 |
| LHX3 | 0.169 | grhprb | -0.064 |
| scrt1a | 0.163 | agxtb | -0.063 |
| XLOC-038699 | 0.160 | scp2a | -0.063 |
| LOC101884740 | 0.160 | abat | -0.062 |
| ebf2 | 0.160 | aqp12 | -0.062 |
| wdr93 | 0.159 | gpd1b | -0.060 |
| foxn4 | 0.157 | si:dkey-16p21.8 | -0.059 |
| LOC103908906 | 0.156 | apoa4b.1 | -0.058 |
| uncx | 0.155 | g6pca.2 | -0.058 |
| phf1 | 0.152 | dpydb | -0.058 |
| chrna5 | 0.152 | pnp4b | -0.058 |
| CABZ01080006.1 | 0.149 | cdo1 | -0.057 |
| BX890591.1 | 0.149 | haao | -0.057 |
| nrcamb | 0.147 | tdo2a | -0.057 |
| gpm6aa | 0.146 | suclg2 | -0.057 |
| BX323820.1 | 0.143 | kng1 | -0.057 |
| scrt2 | 0.142 | chchd10 | -0.057 |
| nrxn3a | 0.141 | mgst1.2 | -0.057 |
| foxg1b | 0.139 | bhmt | -0.056 |
| LOC103909675 | 0.138 | aldh6a1 | -0.056 |
| nhlh2 | 0.137 | gstt1a | -0.056 |
| myt1a | 0.136 | cx32.3 | -0.055 |
| tuba1a | 0.136 | phyhd1 | -0.055 |
| pou2f2a | 0.136 | fabp10a | -0.055 |
| LOC101886822 | 0.133 | etnppl | -0.055 |
| FP101883.1 | 0.132 | ttc36 | -0.055 |
| tuba1c | 0.132 | sult2st2 | -0.055 |