dishevelled-binding antagonist of beta-catenin 1
ZFIN 
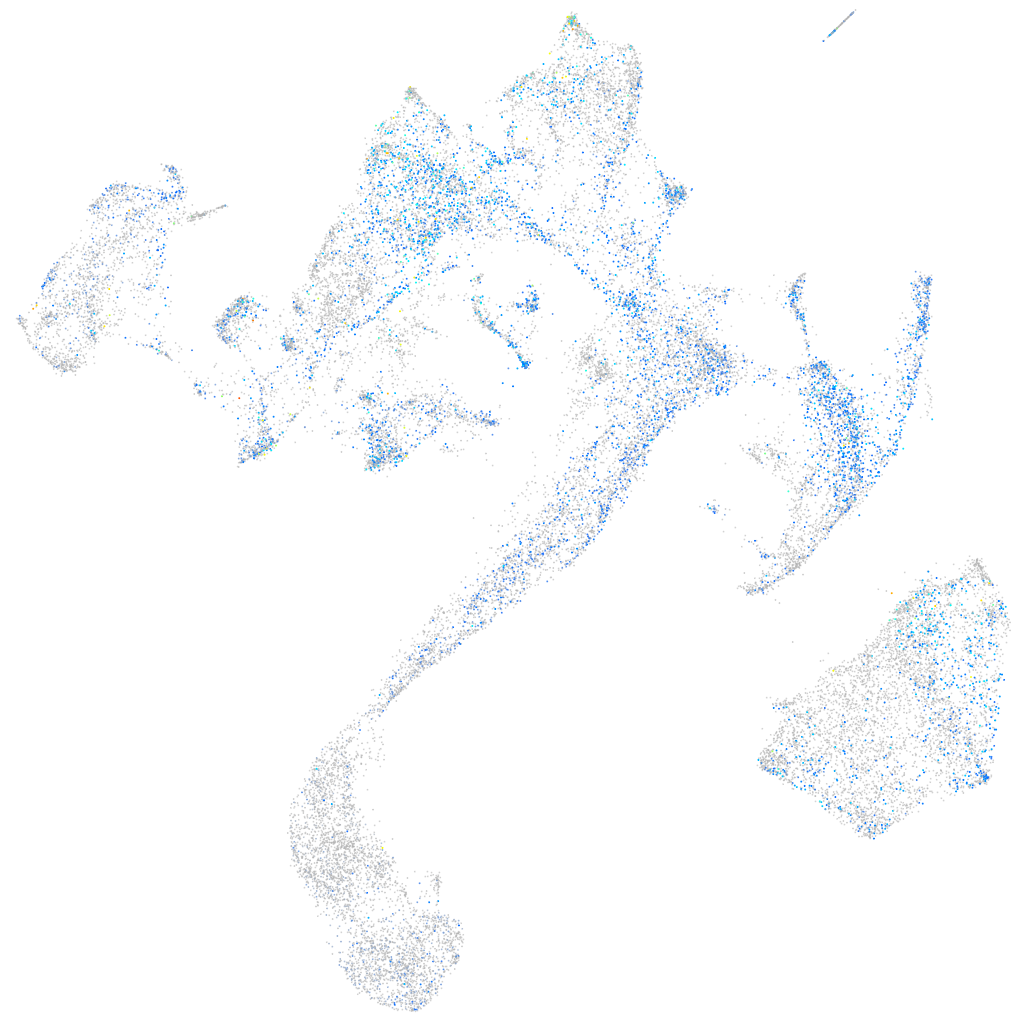
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mfap2 | 0.155 | atp2a1 | -0.125 |
| actb2 | 0.148 | ckmb | -0.124 |
| tcf15 | 0.145 | ckma | -0.124 |
| dmrt2a | 0.145 | gapdh | -0.121 |
| ubc | 0.141 | si:ch73-367p23.2 | -0.120 |
| ppib | 0.140 | tnni2a.4 | -0.119 |
| CR383676.1 | 0.138 | actc1b | -0.118 |
| mdka | 0.137 | XLOC-025819 | -0.118 |
| hsp90ab1 | 0.137 | actn3a | -0.117 |
| zgc:153867 | 0.136 | tnnc2 | -0.117 |
| fstl1b | 0.133 | acta1b | -0.117 |
| meox1 | 0.132 | smyd1a | -0.117 |
| marcksl1a | 0.131 | tnnt3a | -0.117 |
| ssr3 | 0.131 | myoz1b | -0.115 |
| ier5 | 0.131 | mylpfb | -0.115 |
| btg2 | 0.130 | XLOC-001975 | -0.115 |
| id3 | 0.129 | myom1a | -0.115 |
| tspan7 | 0.129 | XLOC-005350 | -0.114 |
| myf5 | 0.128 | aldoab | -0.114 |
| cdh2 | 0.127 | ak1 | -0.114 |
| thbs4b | 0.127 | ldb3b | -0.113 |
| fbn2b | 0.126 | pgam2 | -0.113 |
| sdc4 | 0.126 | XLOC-006515 | -0.113 |
| hspa8 | 0.123 | pabpc4 | -0.112 |
| hnrnpa0l | 0.123 | si:ch211-255p10.3 | -0.112 |
| actb1 | 0.121 | tpma | -0.112 |
| lbx2 | 0.121 | ank1a | -0.112 |
| ier5l | 0.120 | hhatla | -0.111 |
| ism1 | 0.120 | neb | -0.110 |
| boc | 0.119 | myl1 | -0.110 |
| col5a2a | 0.119 | mylz3 | -0.110 |
| fthl27 | 0.119 | mylpfa | -0.109 |
| meis1b | 0.118 | myom2a | -0.108 |
| h3f3c | 0.118 | pvalb2 | -0.108 |
| naca | 0.118 | CABZ01078594.1 | -0.107 |