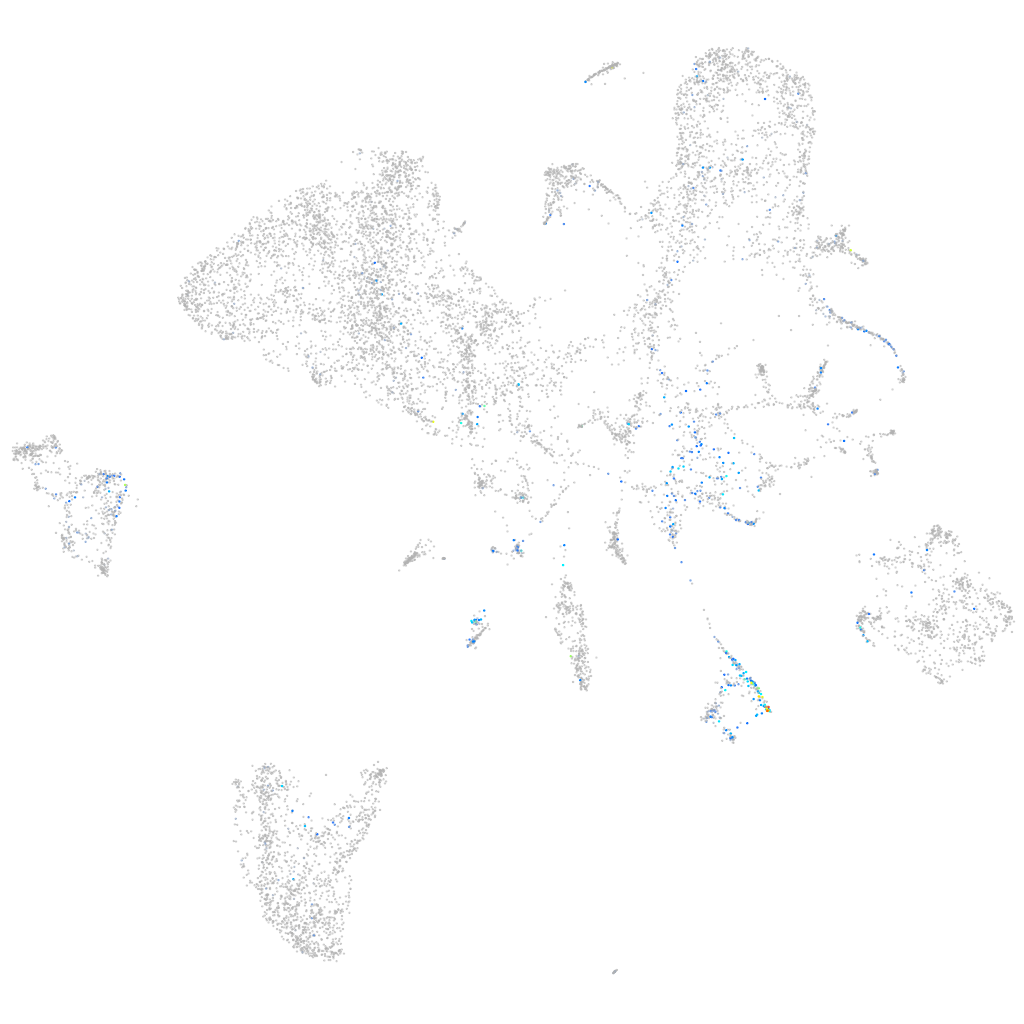
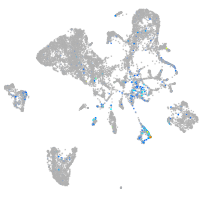
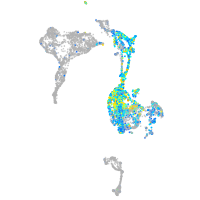
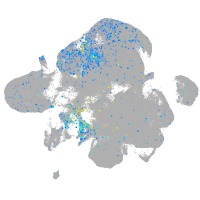
"cysteine-rich, angiogenic inducer, 61 [Source:ZFIN;Acc:ZDB-GENE-040426-3]"
ZFIN 
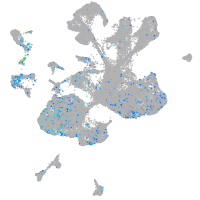
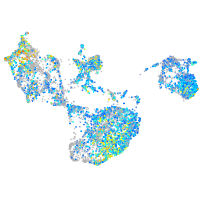
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ctgfa | 0.450 | gapdh | -0.137 |
| CABZ01055365.1 | 0.406 | gatm | -0.129 |
| pnp4a | 0.399 | gamt | -0.127 |
| pmp22b | 0.374 | ahcy | -0.125 |
| tgfb3 | 0.362 | fbp1b | -0.123 |
| sim2.1 | 0.357 | aldob | -0.122 |
| f3a | 0.353 | gpx4a | -0.121 |
| amotl2b | 0.339 | mat1a | -0.118 |
| cd151 | 0.336 | bhmt | -0.115 |
| spns2 | 0.330 | apoc2 | -0.115 |
| cav1 | 0.324 | apoa4b.1 | -0.114 |
| mxra8b | 0.323 | afp4 | -0.112 |
| lama5 | 0.318 | scp2a | -0.111 |
| tekt3 | 0.314 | apoa1b | -0.109 |
| marcksl1a | 0.312 | agxtb | -0.107 |
| edn1 | 0.311 | gstt1a | -0.107 |
| krt94 | 0.308 | pnp4b | -0.106 |
| gna15.1 | 0.308 | grhprb | -0.106 |
| sparc | 0.302 | mdh1aa | -0.103 |
| sim2 | 0.298 | pklr | -0.102 |
| bcam | 0.294 | abat | -0.102 |
| sgms1 | 0.294 | gpd1b | -0.100 |
| sim1b | 0.287 | rdh1 | -0.100 |
| ccdc80 | 0.286 | apoc1 | -0.100 |
| spaca4l | 0.283 | suclg1 | -0.099 |
| col4a5 | 0.280 | tdo2a | -0.098 |
| rnd3b | 0.280 | dap | -0.097 |
| erbb3a | 0.279 | agxta | -0.097 |
| quo | 0.279 | msrb2 | -0.097 |
| vwa1 | 0.278 | suclg2 | -0.096 |
| socs3a | 0.275 | hao1 | -0.096 |
| anxa3b | 0.270 | nipsnap3a | -0.096 |
| col4a6 | 0.266 | aldh6a1 | -0.096 |
| cabp2b | 0.265 | rbp2b | -0.095 |
| cav2 | 0.264 | sult2st2 | -0.095 |