"cytochrome P450, family 2, subfamily AD, polypeptide 3"
ZFIN 
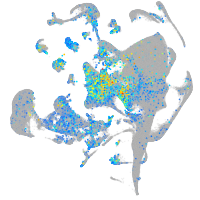
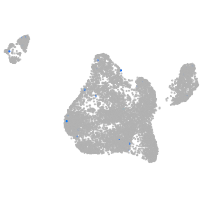
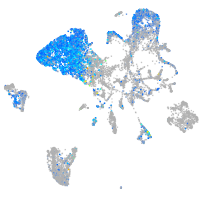
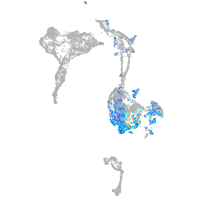
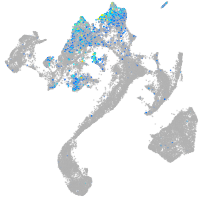
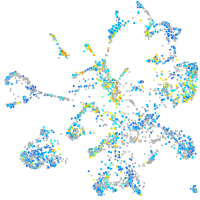
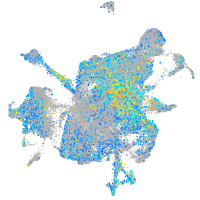
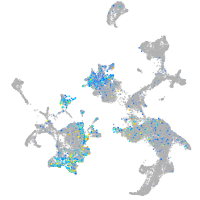
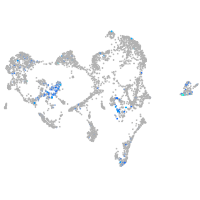
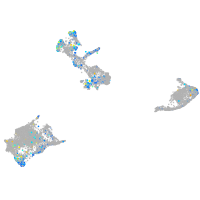
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cyp2n13 | 0.164 | si:ch211-222l21.1 | -0.101 |
| cebpd | 0.163 | ptmab | -0.093 |
| apobb.1 | 0.155 | tubb2b | -0.086 |
| socs3a | 0.151 | hnrnpaba | -0.083 |
| rbp4 | 0.147 | h3f3d | -0.071 |
| apoa4b.1 | 0.147 | khdrbs1a | -0.066 |
| apoa2 | 0.145 | hmgn6 | -0.066 |
| cyp2ad2 | 0.145 | marcksb | -0.063 |
| ces2 | 0.144 | hmgb3a | -0.061 |
| cfhl4 | 0.143 | si:ch211-288g17.3 | -0.061 |
| crip1 | 0.141 | hnrnpa0b | -0.060 |
| col5a1 | 0.141 | cirbpb | -0.059 |
| tfa | 0.141 | hnrnpabb | -0.059 |
| wu:fj16a03 | 0.141 | si:ch73-1a9.3 | -0.059 |
| rdh1 | 0.140 | elavl3 | -0.058 |
| zgc:123103 | 0.139 | sumo3a | -0.057 |
| ttr | 0.139 | nucks1a | -0.057 |
| BX908782.3 | 0.137 | cirbpa | -0.057 |
| serpina1l | 0.137 | marcksl1b | -0.056 |
| apom | 0.137 | hmgb1b | -0.053 |
| serpina1 | 0.137 | h2afvb | -0.052 |
| apoa1b | 0.136 | nova2 | -0.052 |
| fga | 0.135 | ilf2 | -0.052 |
| shbg | 0.132 | tuba1a | -0.052 |
| fgb | 0.131 | tuba1c | -0.052 |
| tmem176 | 0.131 | chd4a | -0.051 |
| cyp3a65 | 0.131 | si:dkey-276j7.1 | -0.051 |
| fabp10a | 0.130 | hnrnpa0a | -0.051 |
| pmp22a | 0.129 | seta | -0.051 |
| lgals2b | 0.128 | zc4h2 | -0.050 |
| col1a2 | 0.128 | setb | -0.050 |
| zgc:112265 | 0.128 | hmga1a | -0.049 |
| comtd1 | 0.128 | hdac1 | -0.049 |
| cfb | 0.127 | tubb5 | -0.048 |
| f2 | 0.126 | hnrnpa1a | -0.048 |