"cytochrome P450, family 1, subfamily C, polypeptide 1"
ZFIN 
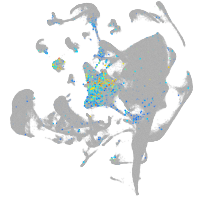
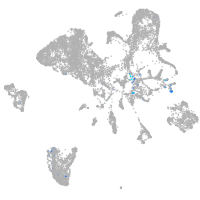
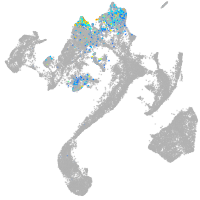
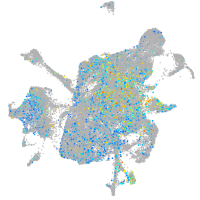
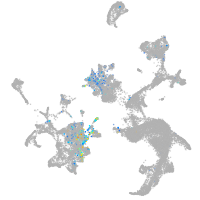
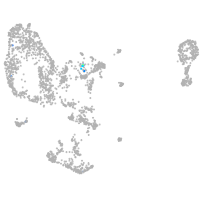
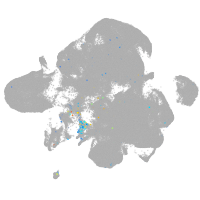
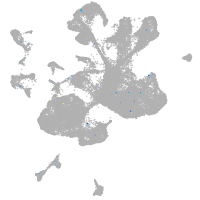
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cyp1c2 | 0.295 | ntn5 | -0.146 |
| mmel1 | 0.241 | si:ch211-62a1.3 | -0.145 |
| si:dkey-57k2.6 | 0.234 | foxf1 | -0.141 |
| gchfr | 0.228 | hlx1 | -0.133 |
| gata5 | 0.228 | nkx2.3 | -0.128 |
| krt94 | 0.223 | csrp1b | -0.124 |
| emid1 | 0.221 | mylkb | -0.122 |
| tnnt2a | 0.220 | foxf2a | -0.113 |
| fbln5 | 0.215 | col4a5 | -0.112 |
| gstm.3 | 0.213 | ckbb | -0.110 |
| twist1b | 0.205 | desmb | -0.109 |
| si:dkey-164f24.2 | 0.202 | col4a6 | -0.108 |
| loxa | 0.202 | cald1b | -0.107 |
| htra1a | 0.199 | tuba8l2 | -0.106 |
| CABZ01092746.1 | 0.198 | XLOC-041870 | -0.106 |
| rbpms2b | 0.198 | BX088707.3 | -0.105 |
| lamb2 | 0.190 | col11a1a | -0.105 |
| lmod2a | 0.188 | ak1 | -0.104 |
| ednraa | 0.187 | inhbaa | -0.101 |
| tbx18 | 0.178 | rbpms2a | -0.100 |
| alpi.1 | 0.178 | BX323087.1 | -0.100 |
| synpo2lb | 0.178 | col6a1 | -0.099 |
| LOC103910903 | 0.178 | ptch2 | -0.098 |
| dub | 0.177 | cnn1b | -0.097 |
| sfrp5 | 0.171 | tpm2 | -0.097 |
| nrp1a | 0.168 | col6a2 | -0.097 |
| lbx2 | 0.168 | tpm1 | -0.097 |
| fhl1b | 0.167 | ndnf | -0.096 |
| lmcd1 | 0.166 | aoc2 | -0.095 |
| smoc1 | 0.166 | acta2 | -0.091 |
| FITM1 (1 of many) | 0.164 | atp5mc1 | -0.090 |
| junba | 0.162 | gapdhs | -0.089 |
| c4b | 0.162 | acta1a | -0.088 |
| creb5b | 0.159 | tagln | -0.088 |
| gsc | 0.159 | hoxc8a | -0.087 |