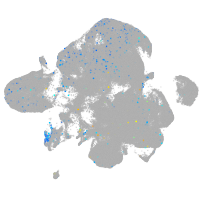
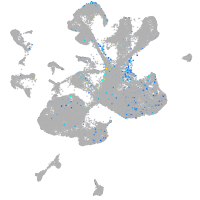
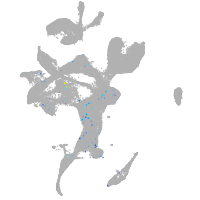
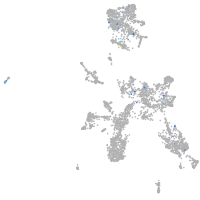
cubilin (intrinsic factor-cobalamin receptor)
ZFIN 
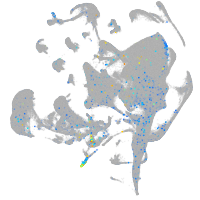
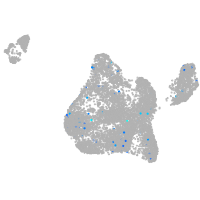
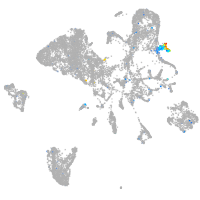
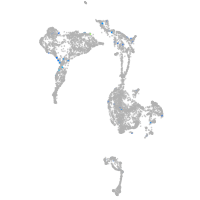
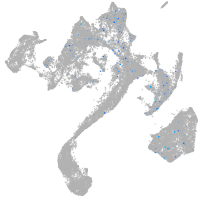
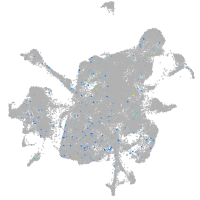
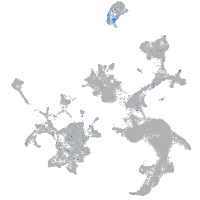
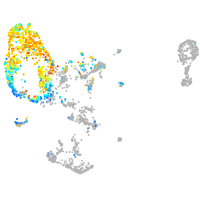
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cart4 | 0.107 | ckbb | -0.054 |
| inka1b | 0.099 | pvalb1 | -0.045 |
| trh | 0.087 | CU634008.1 | -0.044 |
| arid3b | 0.087 | zfhx4 | -0.038 |
| vim | 0.086 | cadm4 | -0.037 |
| tinagl1 | 0.077 | gpm6aa | -0.036 |
| cdx4 | 0.075 | CU467822.1 | -0.035 |
| tuba8l3 | 0.074 | marcksl1b | -0.035 |
| si:ch211-112f3.4 | 0.072 | hbbe1.3 | -0.035 |
| ldlrad2 | 0.070 | pvalb2 | -0.034 |
| cxcr4b | 0.067 | fabp7a | -0.033 |
| lin28a | 0.066 | sncgb | -0.032 |
| CABZ01075068.1 | 0.062 | cspg5a | -0.032 |
| BX001014.2 | 0.061 | slc1a2b | -0.030 |
| LOC108183555 | 0.060 | celf2 | -0.030 |
| greb1l | 0.060 | tnrc6c1 | -0.030 |
| nr6a1a | 0.059 | actc1b | -0.029 |
| cort | 0.057 | snap25a | -0.029 |
| abtb2b | 0.056 | nfil3-5 | -0.028 |
| olig4 | 0.054 | nrxn1a | -0.028 |
| inavaa | 0.054 | elavl4 | -0.028 |
| trim71 | 0.054 | BX664610.1 | -0.028 |
| neurod4 | 0.054 | cplx2 | -0.028 |
| si:dkey-103k4.1 | 0.053 | rbfox1 | -0.028 |
| nes | 0.053 | CR383676.1 | -0.028 |
| dlb | 0.052 | si:ch73-386h18.1 | -0.028 |
| eef1a1l1 | 0.051 | stx1b | -0.027 |
| XLOC-042222 | 0.051 | sypb | -0.027 |
| ebf2 | 0.051 | gpm6ab | -0.027 |
| draxin | 0.051 | hmgb1a | -0.027 |
| hspb1 | 0.050 | si:dkeyp-117h8.2 | -0.026 |
| tp53inp2 | 0.050 | tpma | -0.026 |
| cyb561 | 0.050 | marcksl1a | -0.026 |
| olig2 | 0.049 | slc6a1b | -0.026 |
| cx43.4 | 0.049 | hbae3 | -0.026 |