"cystatin 14b, tandem duplicate 1"
ZFIN 
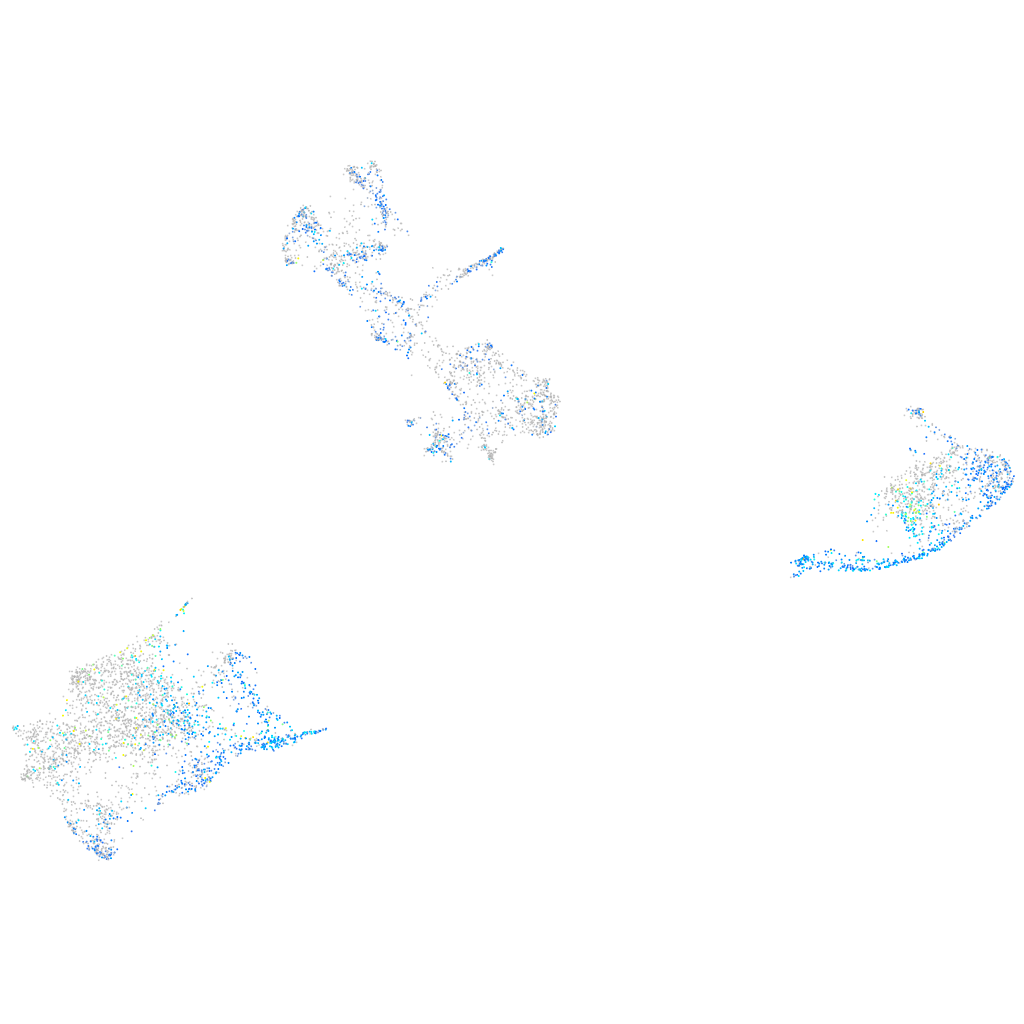
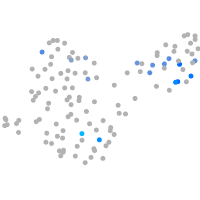
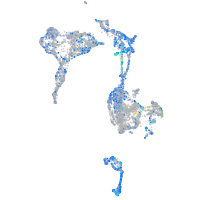
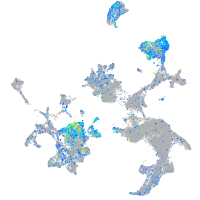
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| qdpra | 0.281 | pvalb2 | -0.160 |
| rab32a | 0.271 | pvalb1 | -0.148 |
| tmem243b | 0.260 | gpm6aa | -0.131 |
| msx1b | 0.255 | ptmaa | -0.126 |
| mitfa | 0.252 | actc1b | -0.122 |
| xbp1 | 0.248 | mdkb | -0.118 |
| tspan36 | 0.244 | stmn1b | -0.115 |
| atp6v1g1 | 0.243 | tuba1c | -0.110 |
| C12orf75 | 0.236 | hbbe1.3 | -0.108 |
| syngr1a | 0.233 | tmsb | -0.106 |
| sec61b | 0.228 | nova2 | -0.106 |
| XLOC-001964 | 0.227 | elavl3 | -0.106 |
| rab38 | 0.227 | hbbe1.1 | -0.105 |
| slc37a2 | 0.225 | mylz3 | -0.105 |
| rnaseka | 0.223 | hbae3 | -0.103 |
| rabl6b | 0.221 | mylpfa | -0.102 |
| tmem243a | 0.218 | tnnt3b | -0.096 |
| naga | 0.217 | epb41a | -0.094 |
| ssr2 | 0.215 | ndrg2 | -0.094 |
| abracl | 0.214 | snap25b | -0.093 |
| ptges3b | 0.213 | hmgb1a | -0.092 |
| ctsba | 0.212 | hmgb1b | -0.090 |
| sypl2b | 0.211 | gng3 | -0.090 |
| pmm2 | 0.211 | sncb | -0.090 |
| gpr143 | 0.210 | elavl4 | -0.088 |
| lman2 | 0.209 | agmo | -0.086 |
| tyr | 0.209 | cadm3 | -0.085 |
| dad1 | 0.209 | mdka | -0.085 |
| ssr3 | 0.209 | apoa2 | -0.084 |
| phlda1 | 0.208 | nme2b.2 | -0.083 |
| pah | 0.208 | CU467822.1 | -0.083 |
| aldoaa | 0.208 | prss59.2 | -0.082 |
| tram1 | 0.207 | atp1a3a | -0.081 |
| zgc:165573 | 0.205 | si:ch211-251b21.1 | -0.080 |
| degs1 | 0.204 | gpr85 | -0.080 |