"crystallin, gamma N2"
ZFIN 
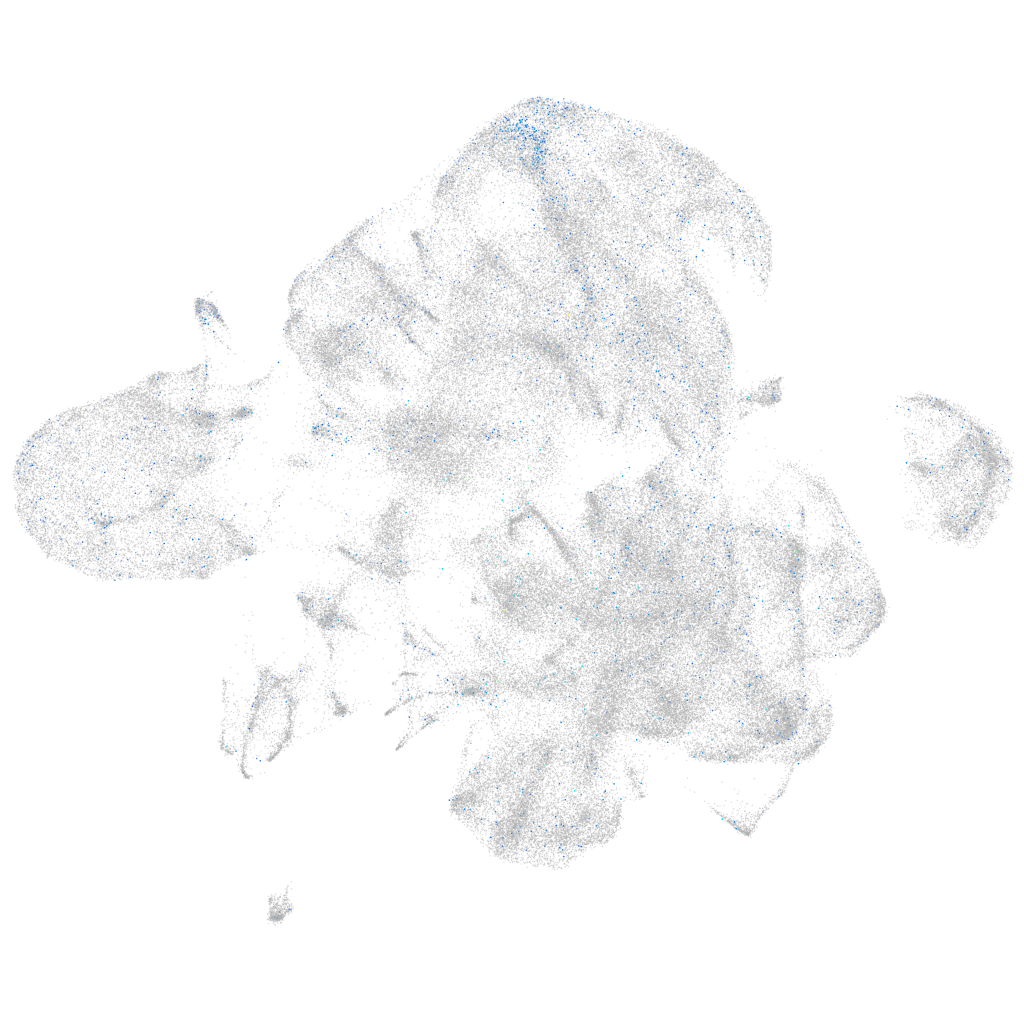
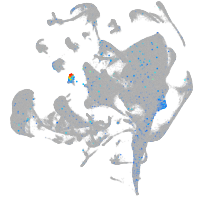
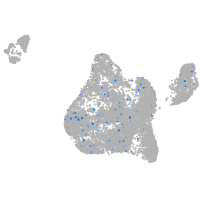
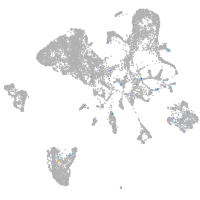
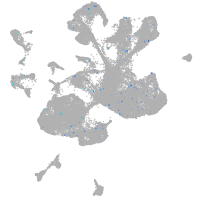
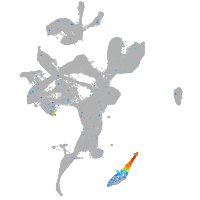
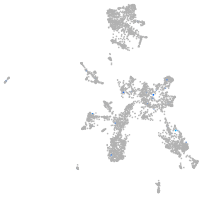
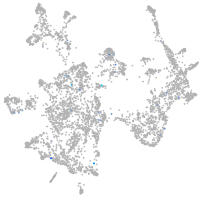
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cryba2b | 0.121 | gpm6aa | -0.026 |
| egr2b | 0.100 | ywhag2 | -0.025 |
| timp2b | 0.095 | gpm6ab | -0.025 |
| mafba | 0.079 | ckbb | -0.025 |
| crygmx | 0.063 | rtn1a | -0.024 |
| cryba4 | 0.063 | stmn1b | -0.024 |
| mpz | 0.062 | gapdhs | -0.024 |
| egr2a | 0.061 | atp6v0cb | -0.023 |
| cyp26c1 | 0.060 | elavl4 | -0.023 |
| crygm3 | 0.058 | myt1la | -0.022 |
| cryba1l1 | 0.054 | sncb | -0.022 |
| lim2.1 | 0.054 | stx1b | -0.022 |
| ackr3b | 0.050 | rbfox1 | -0.021 |
| hoxa3a | 0.049 | rtn1b | -0.021 |
| cxcl12a | 0.049 | si:dkeyp-75h12.5 | -0.021 |
| CABZ01075068.1 | 0.047 | rnasekb | -0.020 |
| dtnba | 0.046 | stxbp1a | -0.020 |
| XLOC-032526 | 0.041 | mt-co2 | -0.019 |
| nppc | 0.040 | tmsb2 | -0.019 |
| XLOC-039121 | 0.040 | sv2a | -0.019 |
| nr2f5 | 0.040 | basp1 | -0.019 |
| ptgdsb.1 | 0.039 | eno2 | -0.019 |
| cldn5a | 0.037 | map4l | -0.018 |
| hoxa2b | 0.037 | COX3 | -0.018 |
| zbtb16b | 0.036 | kdm6bb | -0.018 |
| si:dkey-157g16.6 | 0.036 | map1aa | -0.018 |
| crybb1l2 | 0.036 | snap25a | -0.018 |
| sox3 | 0.035 | tuba2 | -0.018 |
| gas6 | 0.035 | gnao1a | -0.018 |
| eef1a1l1 | 0.035 | nrxn1a | -0.018 |
| pcna | 0.034 | sypb | -0.018 |
| sox21a | 0.034 | atp6v1e1b | -0.018 |
| CR626889.1 | 0.034 | ywhag1 | -0.018 |
| hspb1 | 0.034 | chd5 | -0.018 |
| trim71 | 0.034 | nova1 | -0.018 |