"crystallin, beta A2b"
ZFIN 
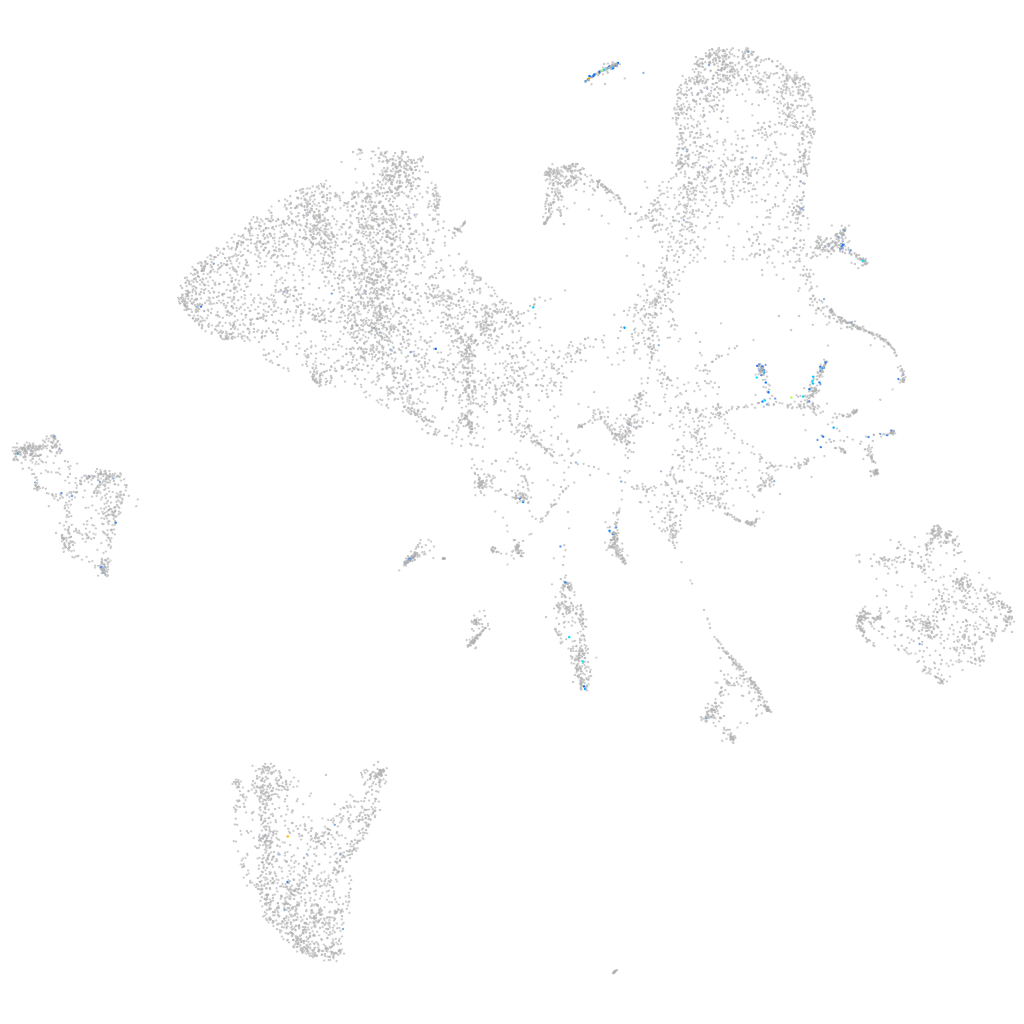
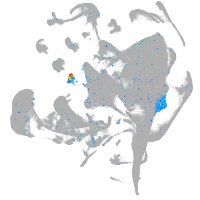
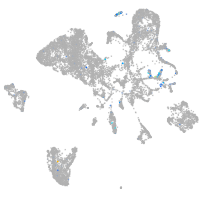
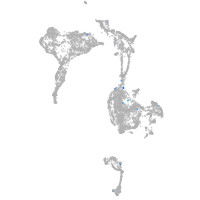
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mipb | 0.259 | aldh7a1 | -0.070 |
| adcyap1a | 0.248 | gamt | -0.069 |
| crygmx | 0.233 | ahcy | -0.069 |
| lrrc34 | 0.233 | rpl6 | -0.068 |
| gja8b | 0.231 | gapdh | -0.068 |
| scg3 | 0.228 | fabp3 | -0.067 |
| fev | 0.225 | mat1a | -0.067 |
| BX255912.1 | 0.213 | gstr | -0.067 |
| egr4 | 0.213 | scp2a | -0.066 |
| mir375-2 | 0.205 | agxta | -0.065 |
| sall1b | 0.203 | si:dkey-16p21.8 | -0.065 |
| lim2.4 | 0.202 | gstt1a | -0.063 |
| kcnk13a | 0.200 | eif4ebp1 | -0.063 |
| insm1b | 0.194 | apoc2 | -0.062 |
| mipa | 0.193 | agxtb | -0.062 |
| pax4 | 0.191 | aldh6a1 | -0.061 |
| penka | 0.186 | gatm | -0.061 |
| trpa1b | 0.185 | cx32.3 | -0.061 |
| nmu | 0.184 | apoa1b | -0.060 |
| cryba4 | 0.183 | pnp4b | -0.060 |
| npy1r | 0.183 | c1qbp | -0.060 |
| neurod1 | 0.182 | apoa4b.1 | -0.060 |
| cx23 | 0.182 | mgst1.2 | -0.060 |
| zgc:101731 | 0.180 | fabp10a | -0.060 |
| mir7a-1 | 0.179 | hspd1 | -0.059 |
| gja8a | 0.179 | ttc36 | -0.059 |
| crybb1l1 | 0.178 | aldh1l1 | -0.059 |
| hepacam2 | 0.177 | gcshb | -0.059 |
| adgrg4b | 0.176 | fth1a | -0.058 |
| si:ch211-261a10.5 | 0.176 | hpda | -0.058 |
| si:dkey-1h6.8 | 0.175 | apoc1 | -0.058 |
| si:ch211-147d7.5 | 0.174 | cpn1 | -0.057 |
| cryba1b | 0.174 | uox | -0.057 |
| lmx1ba | 0.174 | aqp12 | -0.057 |
| c2cd4a | 0.173 | hmgb3b | -0.057 |