"crystallin, beta A2a"
ZFIN 
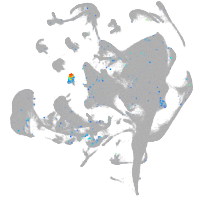
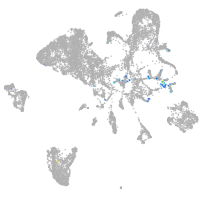
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| insm1a | 0.263 | aldob | -0.095 |
| kcnj11 | 0.259 | eno3 | -0.075 |
| isl1 | 0.259 | gamt | -0.069 |
| pax6b | 0.255 | gapdh | -0.068 |
| mipb | 0.238 | fbp1b | -0.067 |
| neurod1 | 0.234 | glud1b | -0.067 |
| zgc:136683 | 0.230 | aldh7a1 | -0.066 |
| AL954142.1 | 0.227 | ahcy | -0.065 |
| myt1b | 0.226 | aldh6a1 | -0.064 |
| si:ch211-255g12.6 | 0.224 | gstt1a | -0.064 |
| scg2b | 0.219 | cx32.3 | -0.064 |
| scgn | 0.217 | nupr1b | -0.063 |
| gja8b | 0.217 | gstr | -0.062 |
| si:dkey-153k10.9 | 0.217 | hdlbpa | -0.062 |
| pygma | 0.216 | sdr16c5b | -0.061 |
| mir7a-1 | 0.209 | mat1a | -0.061 |
| scg3 | 0.208 | cebpd | -0.061 |
| rprmb | 0.208 | bhmt | -0.060 |
| BX649457.1 | 0.207 | gatm | -0.060 |
| dlb | 0.205 | apoa4b.1 | -0.059 |
| crygmx | 0.204 | si:dkey-16p21.8 | -0.058 |
| kcnk9 | 0.202 | ugt1a7 | -0.057 |
| CR774179.5 | 0.202 | apoa1b | -0.057 |
| si:ch211-213d14.1 | 0.201 | CABZ01032488.1 | -0.057 |
| nkx2.2a | 0.200 | mgst1.2 | -0.057 |
| slc35g2b | 0.199 | aldh1l1 | -0.056 |
| crybb1l1 | 0.198 | rdh1 | -0.056 |
| LOC100537384 | 0.197 | gstp1 | -0.056 |
| ndufa4l2a | 0.197 | ckba | -0.056 |
| cx23 | 0.195 | apoc2 | -0.056 |
| mir375-2 | 0.192 | afp4 | -0.056 |
| rprm3 | 0.190 | agxta | -0.056 |
| gcgb | 0.190 | sult2st2 | -0.056 |
| si:ch211-256e16.4 | 0.190 | scp2a | -0.056 |
| fap | 0.190 | gcshb | -0.055 |