cytokine receptor-like factor 1b
ZFIN 
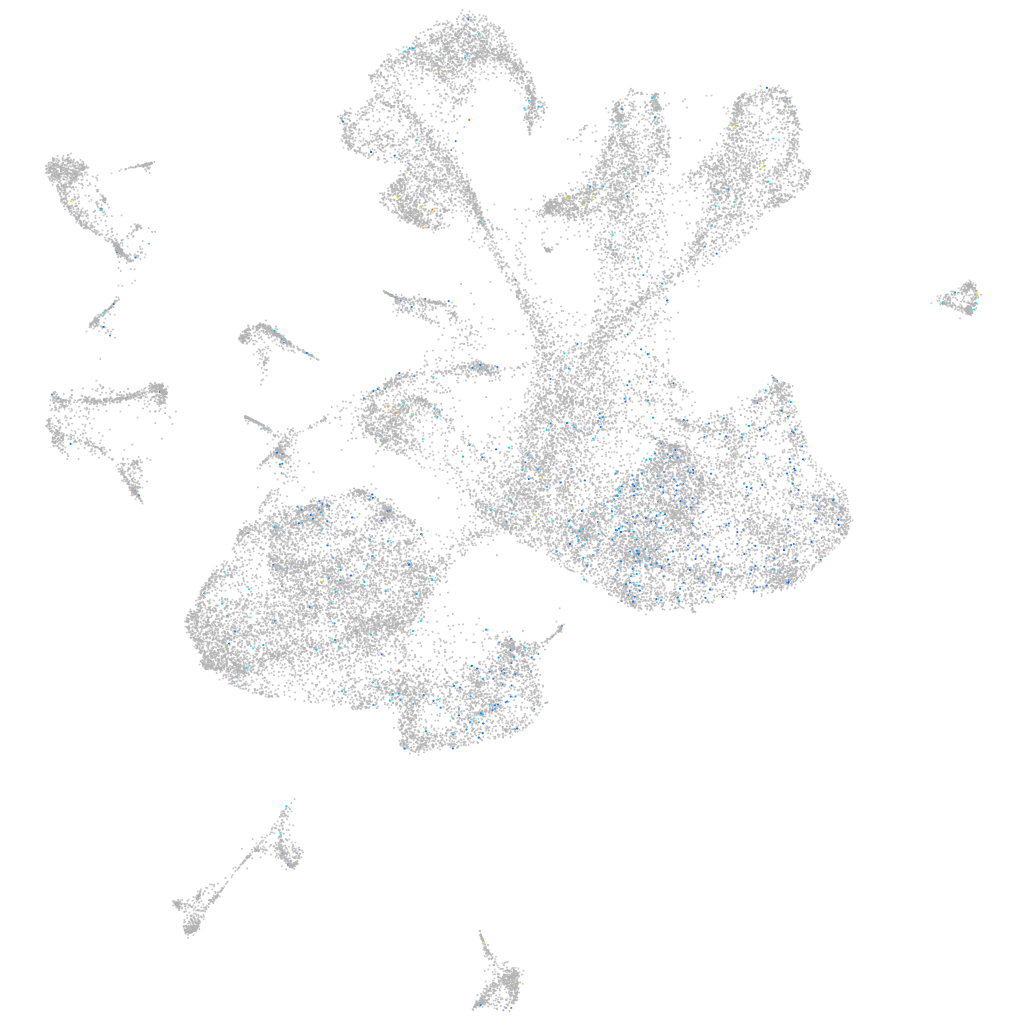
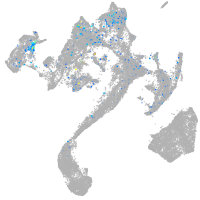
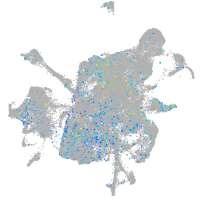
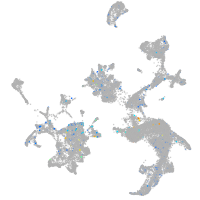
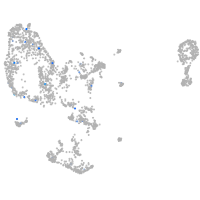
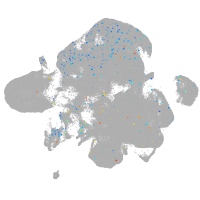
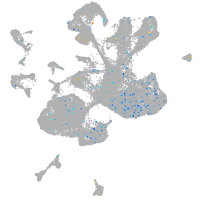
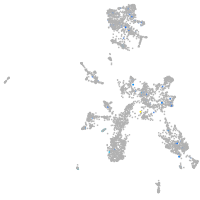
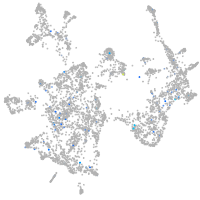
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101883023 | 0.096 | ckbb | -0.033 |
| si:dkey-31g16.2 | 0.068 | rnasekb | -0.032 |
| rrm2 | 0.065 | elavl3 | -0.029 |
| rrm1 | 0.065 | rtn1a | -0.028 |
| si:ch211-111e20.1 | 0.064 | atp6v0cb | -0.027 |
| dek | 0.063 | pvalb2 | -0.027 |
| CR385050.1 | 0.063 | cotl1 | -0.027 |
| pcna | 0.062 | calm1a | -0.027 |
| fbxo5 | 0.062 | h2afx1 | -0.026 |
| zgc:110540 | 0.062 | cadm4 | -0.026 |
| mibp | 0.061 | gap43 | -0.026 |
| stmn1a | 0.060 | myt1b | -0.026 |
| CABZ01005379.1 | 0.059 | atp6v1e1b | -0.025 |
| esco2 | 0.058 | map1aa | -0.025 |
| dut | 0.058 | necap1 | -0.025 |
| chaf1a | 0.058 | syt2a | -0.025 |
| NC-002333.9 | 0.058 | stmn1b | -0.025 |
| LOC108191592 | 0.056 | ppdpfb | -0.025 |
| lbr | 0.056 | elavl4 | -0.025 |
| igfals | 0.055 | gng3 | -0.025 |
| chek1 | 0.054 | si:dkeyp-75h12.5 | -0.024 |
| lig1 | 0.054 | sncb | -0.024 |
| mcm7 | 0.054 | clvs2 | -0.024 |
| LOC101884267 | 0.054 | ywhah | -0.024 |
| LOC101885337 | 0.053 | gpm6ab | -0.024 |
| banf1 | 0.053 | zc4h2 | -0.024 |
| si:dkey-261m9.17 | 0.052 | tuba8l3 | -0.024 |
| anp32b | 0.052 | actc1b | -0.023 |
| unc93a | 0.052 | onecut2 | -0.023 |
| suv39h1b | 0.052 | mllt11 | -0.023 |
| trim35-30 | 0.052 | stxbp1a | -0.023 |
| XLOC-034214 | 0.051 | cplx2 | -0.023 |
| tuba8l4 | 0.051 | gabarapa | -0.023 |
| nasp | 0.051 | CABZ01117348.1 | -0.023 |
| si:dkey-6i22.5 | 0.050 | vamp2 | -0.023 |