"coronin, actin binding protein, 1Ca"
ZFIN 
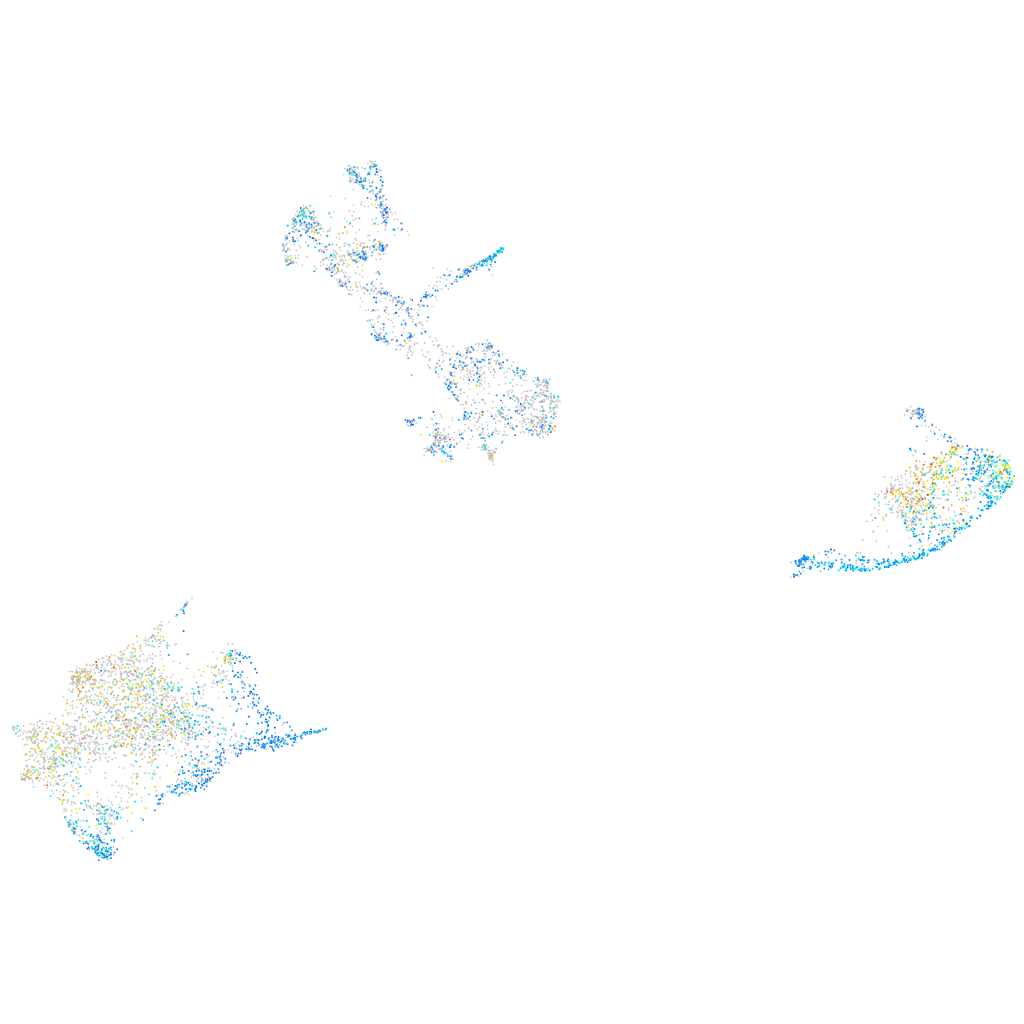
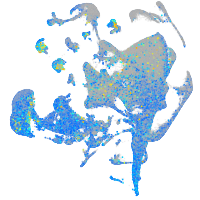
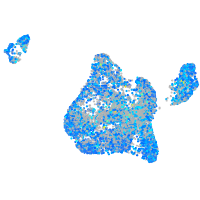
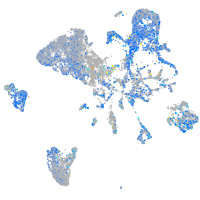
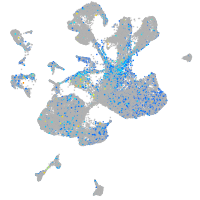
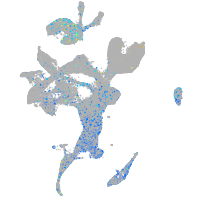
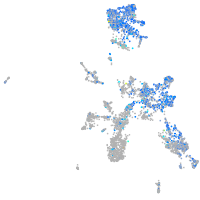
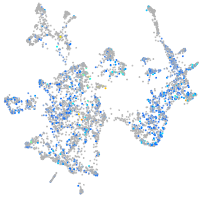
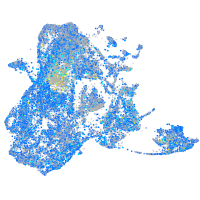
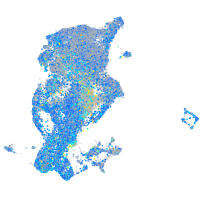
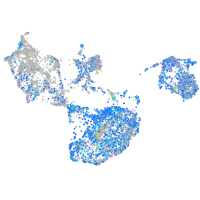
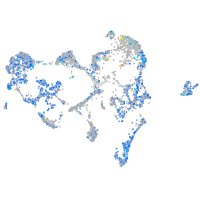
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| oca2 | 0.281 | si:ch211-222l21.1 | -0.144 |
| zgc:91968 | 0.277 | ptmaa | -0.138 |
| kita | 0.268 | hmgn2 | -0.127 |
| pmela | 0.261 | mdkb | -0.125 |
| tyrp1b | 0.251 | si:ch73-1a9.3 | -0.121 |
| prkar1b | 0.250 | nova2 | -0.121 |
| pah | 0.247 | si:dkey-251i10.2 | -0.119 |
| mtbl | 0.246 | CABZ01021592.1 | -0.114 |
| tyrp1a | 0.243 | hmgb2a | -0.108 |
| slc24a5 | 0.241 | elavl3 | -0.108 |
| kcnj13 | 0.240 | hmgb1b | -0.107 |
| vat1 | 0.237 | paics | -0.106 |
| tyr | 0.236 | tuba1c | -0.106 |
| slc39a10 | 0.232 | stmn1a | -0.106 |
| dct | 0.231 | stmn1b | -0.103 |
| lamp1a | 0.226 | gpm6aa | -0.101 |
| si:ch73-389b16.1 | 0.224 | ptmab | -0.099 |
| slc22a2 | 0.224 | tmsb | -0.097 |
| rabl6b | 0.222 | h3f3a | -0.095 |
| slc45a2 | 0.219 | gng3 | -0.095 |
| SPAG9 | 0.218 | h3f3d | -0.092 |
| slc37a2 | 0.218 | gng2 | -0.090 |
| mlpha | 0.216 | zc4h2 | -0.089 |
| hsd20b2 | 0.213 | si:dkey-197i20.6 | -0.089 |
| atp6v0ca | 0.213 | sumo3a | -0.089 |
| spra | 0.211 | fez1 | -0.088 |
| hdlbpa | 0.210 | olfm1b | -0.087 |
| sptan1 | 0.208 | si:ch73-281n10.2 | -0.087 |
| qdpra | 0.206 | sox11a | -0.085 |
| kif5ba | 0.204 | rnasekb | -0.083 |
| rab38 | 0.204 | hmgn7 | -0.083 |
| opn5 | 0.201 | zgc:153426 | -0.083 |
| myo5aa | 0.200 | hmgb3a | -0.082 |
| gstt1a | 0.200 | CU467822.1 | -0.081 |
| tfap2e | 0.194 | apoda.1 | -0.081 |