centriolin
ZFIN 
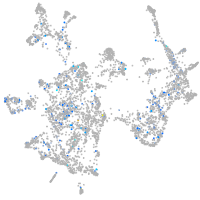
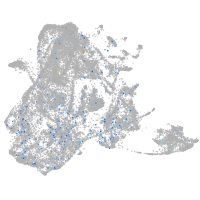
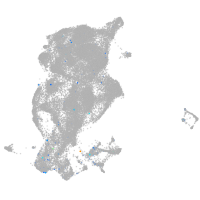
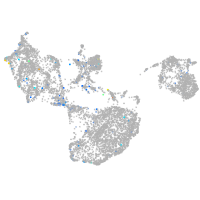
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX511066.2 | 0.280 | si:ch211-139a5.9 | -0.086 |
| XLOC-014193 | 0.269 | gapdh | -0.085 |
| si:dkey-234i14.2 | 0.226 | si:dkeyp-72g9.4 | -0.083 |
| tatdn2 | 0.216 | gstp1 | -0.083 |
| si:ch73-7i4.3 | 0.215 | sod2 | -0.083 |
| tspoap1 | 0.206 | sod1 | -0.083 |
| CABZ01068050.1 | 0.202 | tktb | -0.082 |
| XLOC-019614 | 0.197 | glud1b | -0.079 |
| arl9 | 0.194 | ASS1 | -0.079 |
| CABZ01060030.1 | 0.192 | slc5a12 | -0.078 |
| spaca9 | 0.192 | slc3a2a | -0.078 |
| LOC101886468 | 0.190 | slc7a8a | -0.076 |
| cfap53 | 0.189 | cox7a1 | -0.075 |
| lmnl3 | 0.179 | si:ch211-133l5.7 | -0.075 |
| XLOC-025298 | 0.179 | acy1 | -0.075 |
| fbxo41 | 0.177 | slc34a1a | -0.074 |
| si:dkeyp-34f6.4 | 0.177 | aifm1 | -0.074 |
| armh1 | 0.176 | rgn | -0.074 |
| si:dkey-222p3.1 | 0.171 | gldc | -0.073 |
| cetn4 | 0.171 | enosf1 | -0.073 |
| XLOC-010278 | 0.170 | slc2a11l | -0.073 |
| XLOC-019150 | 0.169 | agxtb | -0.072 |
| fam161a | 0.169 | prdx2 | -0.071 |
| kcna2b | 0.167 | mgst1.2 | -0.071 |
| cnksr2a | 0.167 | slc22a7b.1 | -0.071 |
| myl9b | 0.167 | gstz1 | -0.070 |
| si:ch211-175m2.4 | 0.166 | dpys | -0.070 |
| XLOC-031035 | 0.166 | gpt2l | -0.069 |
| znf1017 | 0.165 | adh8b | -0.069 |
| BX323564.1 | 0.165 | gcshb | -0.068 |
| ccdc13 | 0.164 | cox5b2 | -0.068 |
| pde10a | 0.163 | gstt2 | -0.068 |
| ccdc114 | 0.162 | ifi30 | -0.067 |
| abcd2 | 0.161 | pck2 | -0.067 |
| kiz | 0.161 | amt | -0.067 |