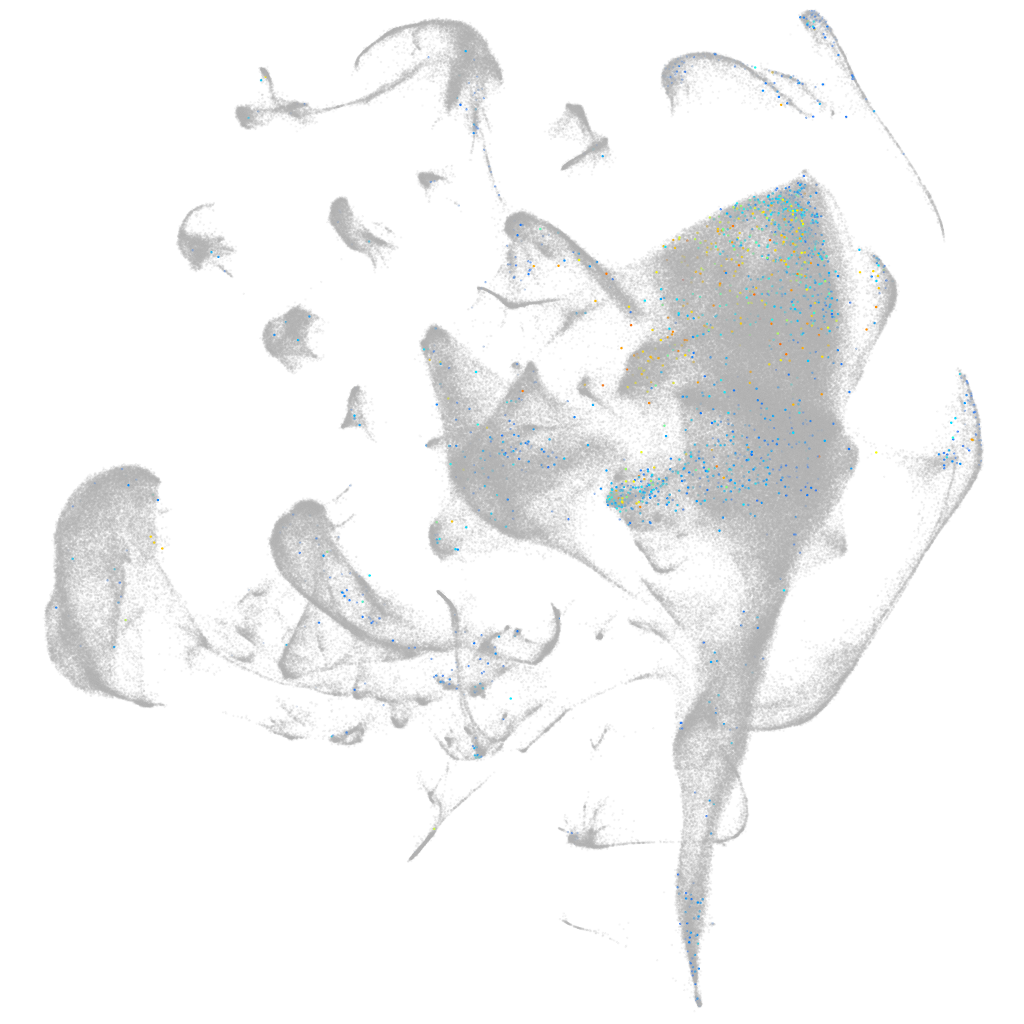connector enhancer of kinase suppressor of Ras 2a
ZFIN 
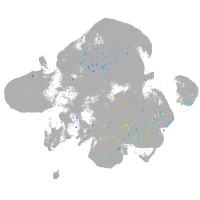
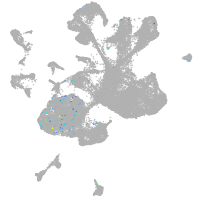
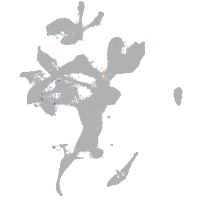
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc1a2b | 0.059 | aldob | -0.022 |
| slc1a3b | 0.057 | wu:fb18f06 | -0.021 |
| si:ch73-305o9.3 | 0.055 | cfl1l | -0.020 |
| camk2a | 0.054 | epcam | -0.020 |
| gpr37l1b | 0.053 | krt4 | -0.020 |
| ywhag2 | 0.053 | apoc1 | -0.019 |
| slc6a11b | 0.052 | apoeb | -0.019 |
| slc6a1b | 0.052 | cyt1l | -0.019 |
| CR936968.2 | 0.052 | lye | -0.019 |
| cspg5a | 0.051 | ppl | -0.019 |
| gria2b | 0.050 | agr1 | -0.018 |
| zgc:153426 | 0.050 | anxa1c | -0.018 |
| ndrg3a | 0.049 | cd9b | -0.018 |
| vsnl1b | 0.049 | cdh1 | -0.018 |
| zgc:65894 | 0.049 | f11r.1 | -0.018 |
| eno2 | 0.048 | gsnb | -0.018 |
| gnao1a | 0.048 | si:ch211-105c13.3 | -0.018 |
| map1aa | 0.048 | si:ch211-195b11.3 | -0.018 |
| map1ab | 0.048 | spint2 | -0.018 |
| ptprz1b | 0.048 | tcima | -0.018 |
| ywhag1 | 0.048 | zgc:153284 | -0.018 |
| gapdhs | 0.047 | zgc:193505 | -0.018 |
| NPAS3 | 0.047 | aep1 | -0.017 |
| atp1a1b | 0.046 | ahnak | -0.017 |
| atp1b4 | 0.046 | anxa1a | -0.017 |
| cdk5r2a | 0.046 | BX927258.1 | -0.017 |
| grm5a | 0.046 | cast | -0.017 |
| negr1 | 0.046 | ckap4 | -0.017 |
| prkcg | 0.046 | cldne | -0.017 |
| slc6a1a | 0.046 | cx43.4 | -0.017 |
| sv2a | 0.046 | cyt1 | -0.017 |
| calm3a | 0.046 | evpla | -0.017 |
| aldocb | 0.045 | hdlbpa | -0.017 |
| stmn2a | 0.045 | icn2 | -0.017 |
| ca4a | 0.044 | krt5 | -0.017 |