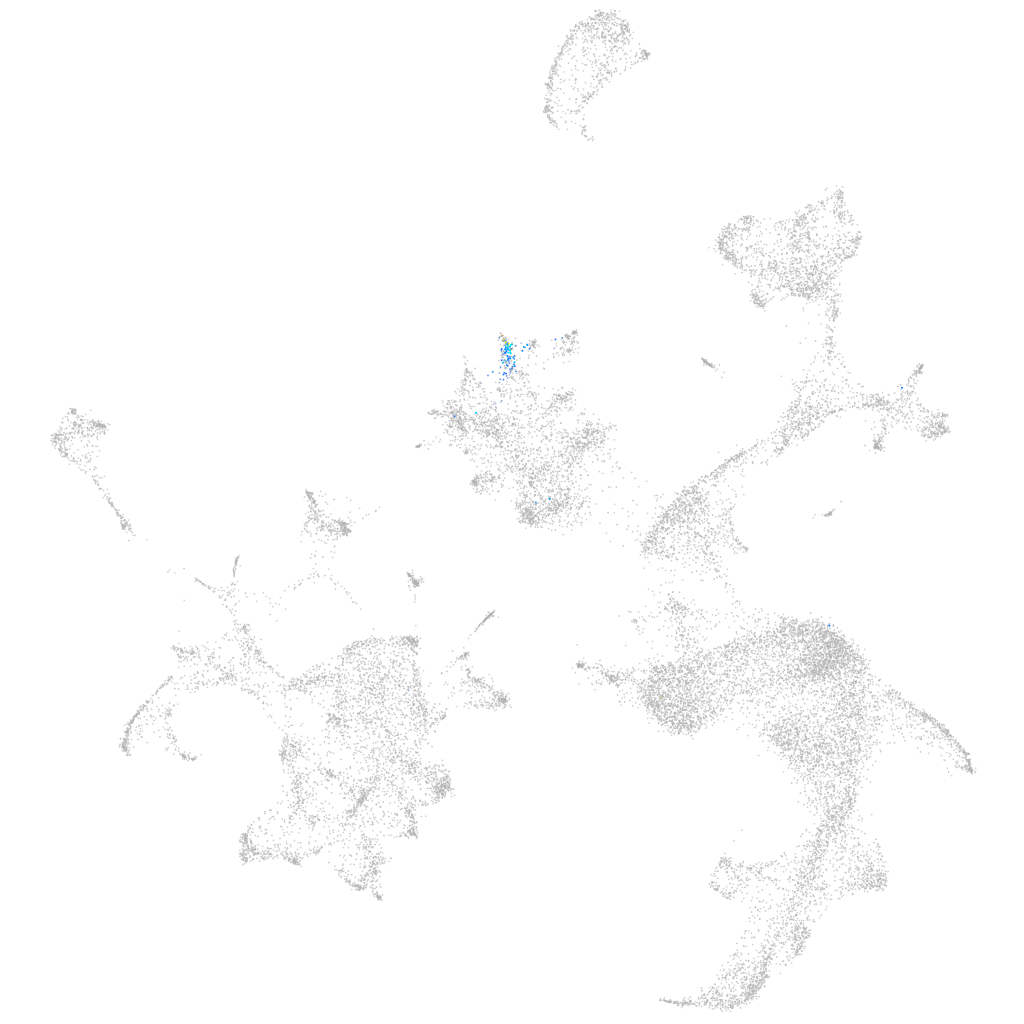
cyclic nucleotide gated channel subunit alpha 3b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prph2a | 0.553 | rpl9 | -0.038 |
| pdcb | 0.534 | zgc:56493 | -0.032 |
| rgs9a | 0.531 | hbbe3 | -0.029 |
| guk1b | 0.515 | rps12 | -0.028 |
| cngb3.2 | 0.497 | msna | -0.027 |
| grk7a | 0.491 | pfn1 | -0.026 |
| rcvrn2 | 0.490 | arhgdia | -0.026 |
| si:ch1073-469d17.2 | 0.490 | urod | -0.025 |
| pde6c | 0.486 | cdh5 | -0.025 |
| arr3a | 0.484 | nutf2l | -0.024 |
| slc25a3a | 0.482 | mob1a | -0.024 |
| gngt2a | 0.482 | clec14a | -0.024 |
| si:ch211-285j22.3 | 0.481 | si:dkey-112e7.2 | -0.024 |
| gnat2 | 0.475 | etv2 | -0.023 |
| zgc:73359 | 0.473 | rpsa | -0.023 |
| gnb3b | 0.470 | rhocb | -0.023 |
| si:dkey-17e16.15 | 0.462 | yrk | -0.022 |
| cnga3a | 0.458 | srgn | -0.022 |
| gabra6a | 0.454 | tspan4b | -0.022 |
| tmx3a | 0.453 | rplp2 | -0.022 |
| arl3l2 | 0.450 | hhex | -0.022 |
| rcvrn3 | 0.449 | she | -0.022 |
| si:ch211-81a5.8 | 0.448 | hmbsa | -0.022 |
| arl3l1 | 0.444 | si:dkeyp-97a10.2 | -0.022 |
| ckmt2a | 0.436 | sox7 | -0.022 |
| sv2ba | 0.429 | myct1a | -0.022 |
| rbp4l | 0.428 | tagln2 | -0.022 |
| grk1b | 0.425 | ucp2 | -0.021 |
| gucy2d | 0.423 | plk2b | -0.021 |
| elovl4b | 0.414 | ecscr | -0.021 |
| rcvrna | 0.412 | hmbsb | -0.021 |
| ldhbb | 0.410 | tie1 | -0.021 |
| si:dkey-220f10.4 | 0.409 | egfl7 | -0.021 |
| tmem237a | 0.408 | ramp2 | -0.021 |
| atp2b1b | 0.408 | eef1a1l1 | -0.021 |