"CLN3 lysosomal/endosomal transmembrane protein, battenin"
ZFIN 
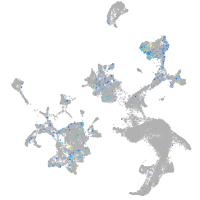
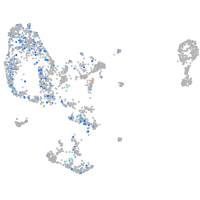
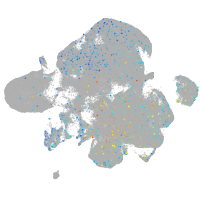
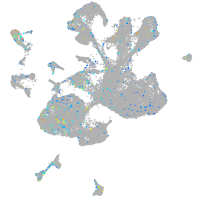
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rnasekb | 0.051 | hmga1a | -0.047 |
| gapdhs | 0.050 | hmgb2a | -0.046 |
| si:dkey-33c3.1 | 0.049 | si:ch211-222l21.1 | -0.039 |
| atp6v1e1b | 0.048 | npm1a | -0.039 |
| CR383676.1 | 0.047 | nop58 | -0.038 |
| atp6v0cb | 0.047 | hmgb2b | -0.038 |
| atp6ap2 | 0.046 | dkc1 | -0.038 |
| aldocb | 0.045 | ncl | -0.038 |
| atpv0e2 | 0.044 | fbl | -0.037 |
| si:dkey-208m12.3 | 0.044 | rps12 | -0.037 |
| rtn1a | 0.044 | hspb1 | -0.036 |
| scg2b | 0.044 | si:ch73-281n10.2 | -0.036 |
| cpe | 0.043 | si:ch211-152c2.3 | -0.036 |
| si:dkeyp-75h12.5 | 0.042 | mki67 | -0.035 |
| calm1a | 0.042 | cirbpa | -0.035 |
| atp6v1g1 | 0.041 | hnrnpa1b | -0.034 |
| atp6v1h | 0.041 | snrpb | -0.034 |
| sncb | 0.041 | nop2 | -0.034 |
| ywhag2 | 0.040 | seta | -0.033 |
| ppdpfb | 0.040 | nop56 | -0.033 |
| eno1a | 0.039 | lig1 | -0.033 |
| egr4 | 0.039 | ranbp1 | -0.033 |
| sncgb | 0.039 | ddx18 | -0.032 |
| calm3a | 0.039 | top1l | -0.032 |
| ndrg3a | 0.038 | ube2c | -0.032 |
| calm1b | 0.038 | tuba8l4 | -0.032 |
| stmn2a | 0.038 | rpl29 | -0.032 |
| sh3gl2a | 0.038 | snu13b | -0.032 |
| vamp2 | 0.038 | stm | -0.031 |
| sypb | 0.038 | rsl1d1 | -0.031 |
| zgc:65894 | 0.037 | chaf1a | -0.031 |
| atp6v0b | 0.037 | si:dkey-151g10.6 | -0.031 |
| map1aa | 0.037 | nasp | -0.031 |
| ccni | 0.037 | cbx3a | -0.031 |
| tpi1b | 0.037 | nhp2 | -0.031 |