calmegin
ZFIN 
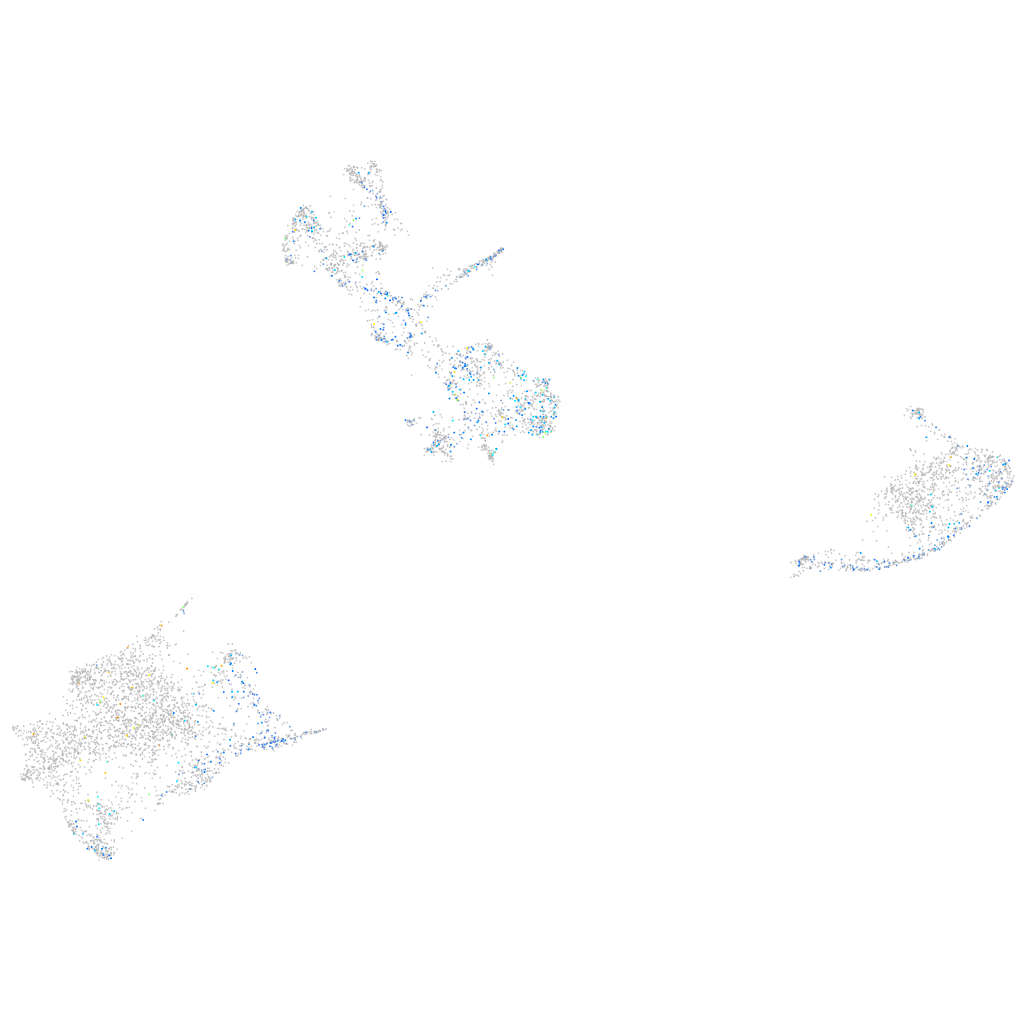
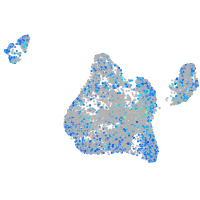
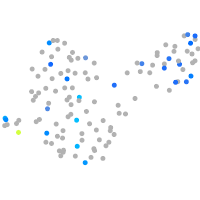
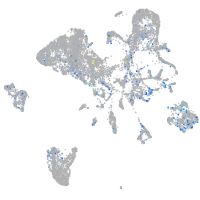
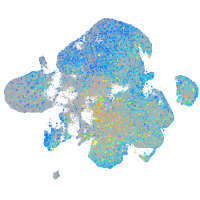
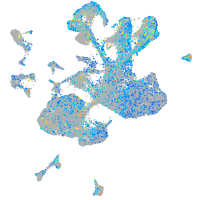
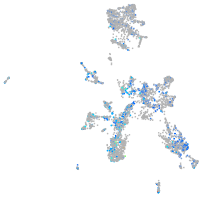
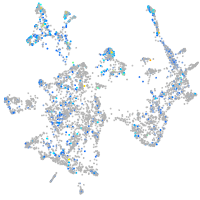
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chd4a | 0.146 | zgc:110339 | -0.095 |
| rtn1a | 0.145 | gstp1 | -0.095 |
| lgi1a | 0.145 | si:dkey-251i10.2 | -0.079 |
| hmgb1b | 0.141 | gch2 | -0.076 |
| hnrnpa0a | 0.141 | uraha | -0.074 |
| nova2 | 0.140 | CABZ01021592.1 | -0.073 |
| rnasekb | 0.139 | slc22a7a | -0.071 |
| pdia6 | 0.139 | pts | -0.067 |
| hnrnpa0b | 0.136 | aqp3a | -0.066 |
| syncrip | 0.135 | bscl2l | -0.064 |
| gpm6aa | 0.135 | pvalb2 | -0.063 |
| ribc1 | 0.135 | mdh1aa | -0.060 |
| wtap | 0.134 | si:ch211-251b21.1 | -0.058 |
| tubb2b | 0.134 | akr1b1 | -0.057 |
| hnrnpaba | 0.134 | krt18b | -0.054 |
| fam168a | 0.133 | actb2 | -0.053 |
| cadm3 | 0.132 | tmem130 | -0.053 |
| tmeff1b | 0.132 | prdx5 | -0.052 |
| seta | 0.132 | krt8 | -0.049 |
| si:ch211-288g17.3 | 0.131 | bco1 | -0.048 |
| atp2b3a | 0.130 | agmo | -0.047 |
| ilf2 | 0.127 | aox5 | -0.047 |
| tmpob | 0.127 | pvalb1 | -0.046 |
| rbm8a | 0.127 | gyg1b | -0.046 |
| atp1a3a | 0.127 | gpd1b | -0.045 |
| calr | 0.127 | aldob | -0.044 |
| lsm7 | 0.127 | si:dkey-151g10.6 | -0.044 |
| tubb5 | 0.126 | actc1b | -0.043 |
| atp6v0cb | 0.126 | pfn1 | -0.043 |
| hmgn2 | 0.125 | nupr1a | -0.040 |
| epb41a | 0.124 | sult1st1 | -0.040 |
| si:ch73-1a9.3 | 0.123 | csrp2 | -0.040 |
| marcksb | 0.123 | zgc:92161 | -0.039 |
| ankrd12 | 0.123 | cmbl | -0.039 |
| rbbp4 | 0.123 | si:zfos-1069f5.1 | -0.038 |