cell death inducing DFFA like effector a
ZFIN 
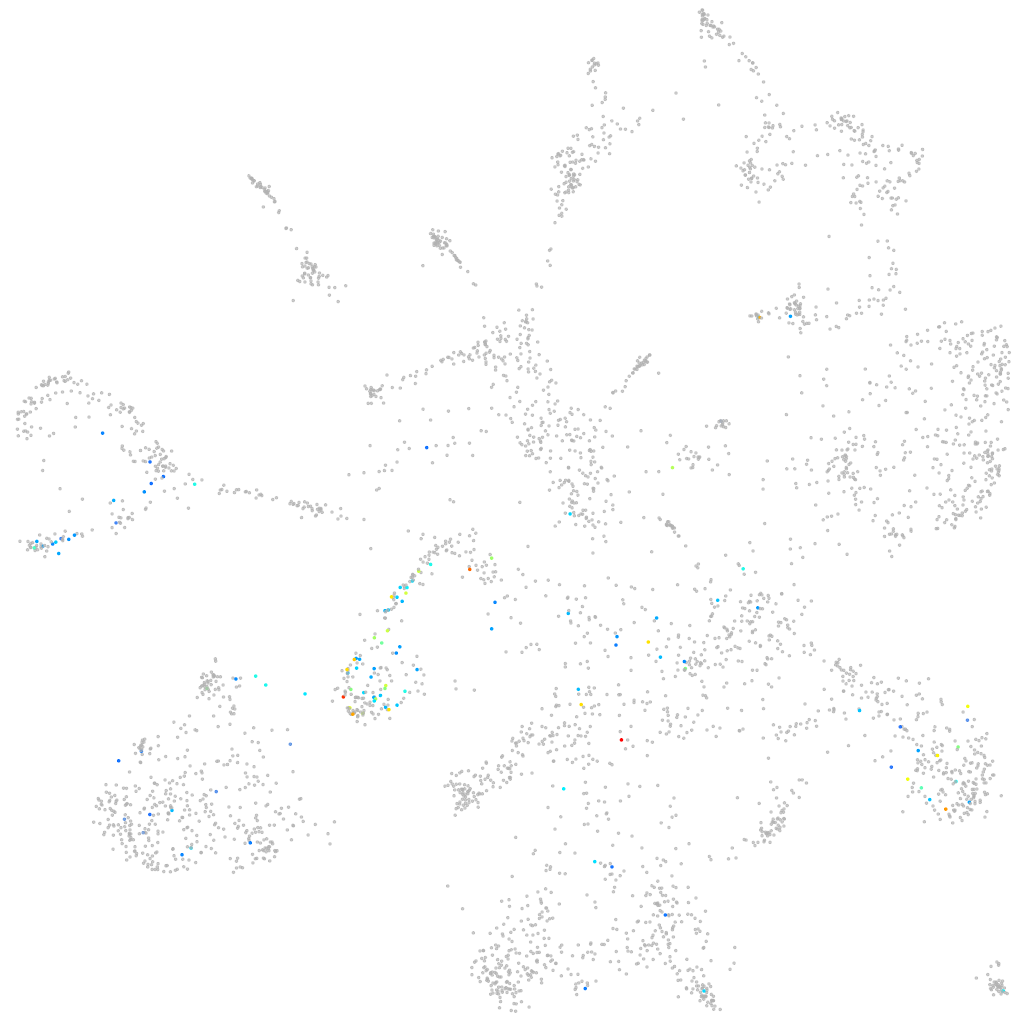
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nrg1 | 0.280 | foxp4 | -0.095 |
| foxl1 | 0.263 | cfd | -0.093 |
| CR318588.3 | 0.261 | krt8 | -0.093 |
| ptgs2a | 0.239 | cavin1b | -0.084 |
| vmo1b | 0.231 | hand2 | -0.079 |
| plk2a | 0.228 | podxl | -0.078 |
| col4a5 | 0.217 | cav1 | -0.075 |
| si:ch211-226m16.2 | 0.216 | cavin2b | -0.074 |
| col4a6 | 0.216 | cdon | -0.072 |
| zgc:195173 | 0.211 | postnb | -0.071 |
| XLOC-040517 | 0.207 | jam2b | -0.070 |
| bmp2b | 0.203 | rhag | -0.069 |
| bgnb | 0.202 | nrp1a | -0.068 |
| dcn | 0.195 | rnd3b | -0.068 |
| ptger2b | 0.193 | cd151 | -0.067 |
| ecrg4a | 0.183 | cdh2 | -0.066 |
| laynb | 0.179 | rplp2 | -0.065 |
| efnb1 | 0.178 | krt18a.1 | -0.065 |
| si:ch211-62a1.3 | 0.178 | ftr82 | -0.064 |
| mgll | 0.176 | slit3 | -0.063 |
| srpx | 0.176 | ephb3 | -0.062 |
| bmp3 | 0.174 | mdka | -0.061 |
| rasgrp2l | 0.174 | lurap1 | -0.061 |
| bmp16 | 0.174 | kank2 | -0.060 |
| tpst2 | 0.173 | krt91 | -0.059 |
| bmp8a | 0.172 | tmem88b | -0.059 |
| vwde | 0.170 | tpm4b | -0.058 |
| LOC108180013 | 0.170 | vat1 | -0.057 |
| col6a2 | 0.168 | pcolcea | -0.057 |
| LOC101885558 | 0.165 | hlx1 | -0.057 |
| rab25a | 0.165 | gas1a | -0.057 |
| gulp1b | 0.162 | lsp1 | -0.056 |
| foxf2b | 0.159 | boc | -0.056 |
| adamtsl5 | 0.159 | tmem88a | -0.056 |
| sema3bl | 0.158 | desmb | -0.056 |