carbohydrate (chondroitin 6) sulfotransferase 3a
ZFIN 
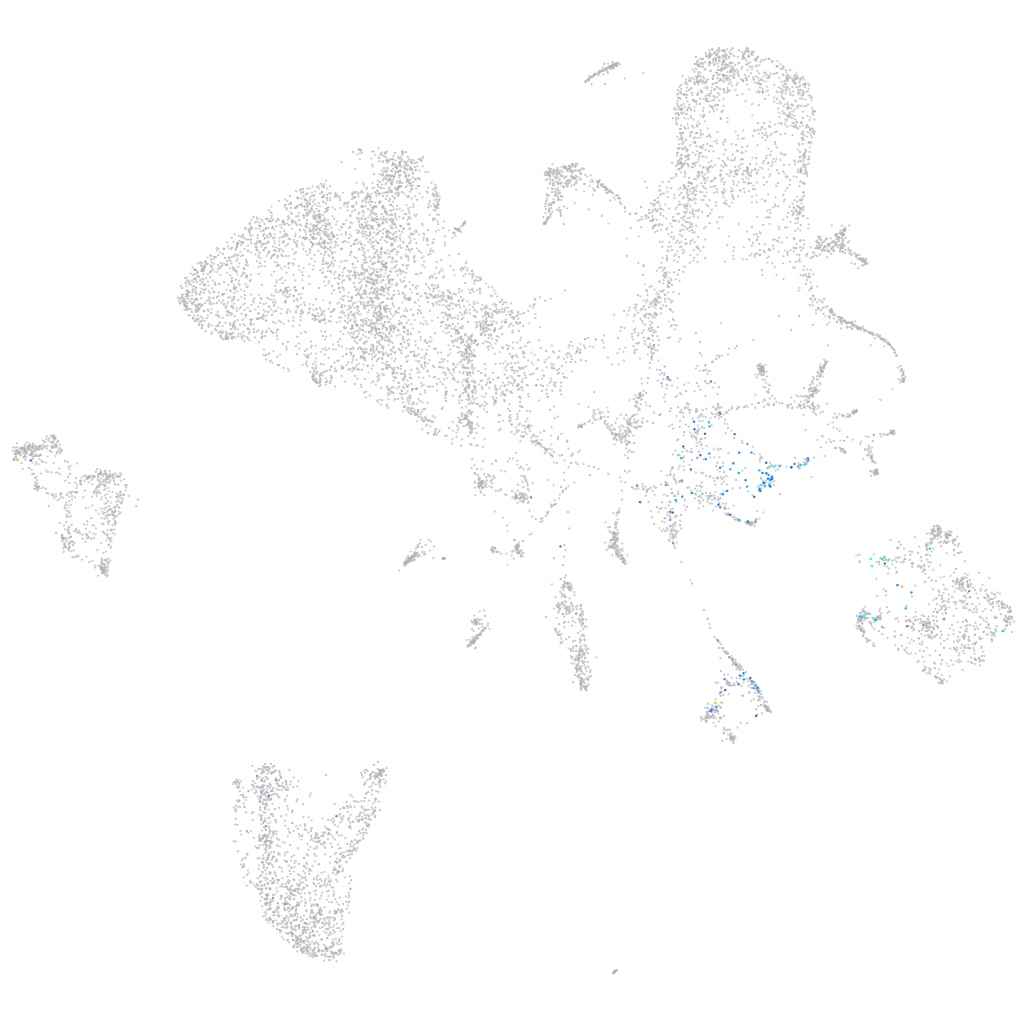
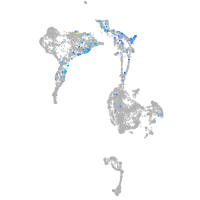
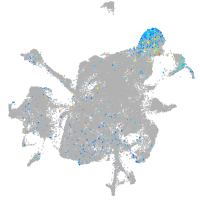
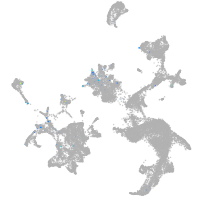
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hapln1b | 0.306 | gapdh | -0.125 |
| ndnf | 0.298 | aldob | -0.122 |
| fgfrl1b | 0.278 | ahcy | -0.119 |
| emid1 | 0.265 | eno3 | -0.117 |
| fstl1a | 0.256 | sod1 | -0.114 |
| vcana | 0.255 | gamt | -0.100 |
| lsp1 | 0.249 | atp5if1b | -0.100 |
| col8a1a | 0.248 | fbp1b | -0.099 |
| gdf6a | 0.236 | apoa1b | -0.098 |
| bcam | 0.231 | gpx4a | -0.097 |
| tpm4a | 0.231 | tpi1b | -0.097 |
| fgf10b | 0.229 | pklr | -0.097 |
| sall4 | 0.227 | apoa4b.1 | -0.097 |
| plod2 | 0.226 | apoc2 | -0.096 |
| gdf6b | 0.226 | sod2 | -0.096 |
| pltp | 0.224 | gatm | -0.095 |
| hhip | 0.224 | lgals2b | -0.095 |
| apela | 0.218 | scp2a | -0.094 |
| CABZ01075068.1 | 0.217 | gstt1a | -0.093 |
| lin28a | 0.214 | afp4 | -0.092 |
| crtap | 0.214 | eef1da | -0.091 |
| raraa | 0.214 | mdh1aa | -0.090 |
| ptgs1 | 0.213 | suclg1 | -0.090 |
| oc90 | 0.210 | atp5mc1 | -0.088 |
| BX001014.2 | 0.208 | ugt1a7 | -0.086 |
| XLOC-034373 | 0.206 | bhmt | -0.086 |
| wnt2bb | 0.206 | apoa2 | -0.086 |
| fkbp9 | 0.205 | BX908782.3 | -0.086 |
| fbn2b | 0.205 | pgk1 | -0.086 |
| nrp2b | 0.203 | mat1a | -0.086 |
| tmem108 | 0.203 | aldh6a1 | -0.085 |
| pcdh19 | 0.197 | glud1b | -0.085 |
| efna5a | 0.197 | gcshb | -0.084 |
| lamb1a | 0.195 | sult2st2 | -0.083 |
| cdx4 | 0.194 | clic5b | -0.083 |