"cholinergic receptor, nicotinic, alpha 3"
ZFIN 
Other cell groups


all cells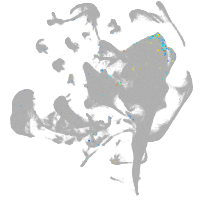 |
blastomeres |
PGCs |
endoderm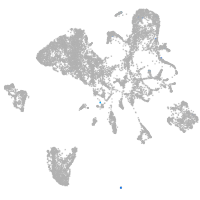 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 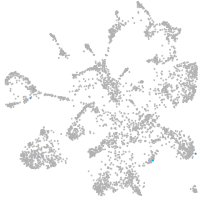 |
mesenchyme 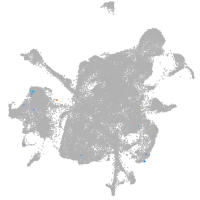 |
hematopoietic / vasculature  |
pronephros  |
neural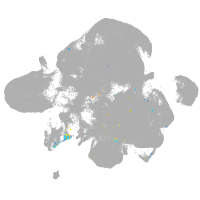 |
spinal cord / glia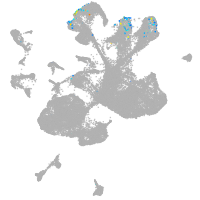 |
eye |
taste / olfactory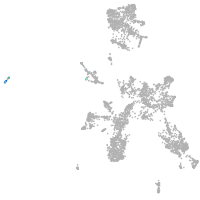 |
otic / lateral line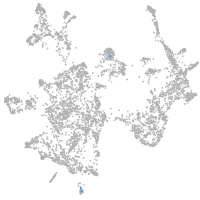 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chrnb4 | 0.284 | aldob | -0.024 |
| vipb | 0.164 | cx43.4 | -0.020 |
| FO907089.1 | 0.131 | hspb1 | -0.019 |
| phox2a | 0.118 | lig1 | -0.018 |
| slc18a3a | 0.113 | mki67 | -0.018 |
| chata | 0.099 | apoeb | -0.017 |
| slc5a7a | 0.099 | banf1 | -0.017 |
| vip | 0.099 | cast | -0.017 |
| pcsk1nl | 0.094 | ccnd1 | -0.017 |
| phox2bb | 0.094 | chaf1a | -0.017 |
| si:dkey-85n7.6 | 0.094 | dkc1 | -0.017 |
| si:ch211-202h22.9 | 0.094 | eif4ebp3l | -0.017 |
| cplx2 | 0.093 | id1 | -0.017 |
| sv2c | 0.092 | nop58 | -0.017 |
| hand2 | 0.091 | polr3gla | -0.017 |
| trarg1a | 0.091 | pou5f3 | -0.017 |
| chrna5 | 0.090 | si:ch211-152c2.3 | -0.017 |
| ret | 0.090 | stm | -0.017 |
| cplx2l | 0.089 | asb11 | -0.016 |
| nefma | 0.089 | epcam | -0.016 |
| anxa13l | 0.087 | fbl | -0.016 |
| atp2b3a | 0.086 | fbxo5 | -0.016 |
| xgb | 0.084 | hdlbpa | -0.016 |
| l1cama | 0.083 | nop56 | -0.016 |
| stmn2a | 0.083 | npm1a | -0.016 |
| tspan2b | 0.083 | sdc4 | -0.016 |
| ribc1 | 0.081 | si:dkey-16p21.8 | -0.016 |
| amigo3 | 0.080 | si:dkey-66i24.9 | -0.016 |
| prph | 0.080 | tpx2 | -0.016 |
| stmn2b | 0.080 | zgc:110425 | -0.016 |
| tmie | 0.080 | zgc:56699 | -0.016 |
| stmn3 | 0.079 | apoc1 | -0.015 |
| LOC100537369 | 0.079 | arf1 | -0.015 |
| ank2a | 0.078 | asph | -0.015 |
| gap43 | 0.077 | ccna2 | -0.015 |




