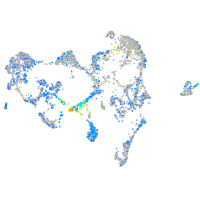
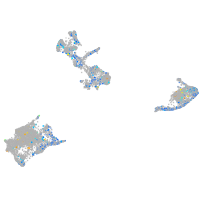
calcineurin-like EF-hand protein 1
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| loxl5b | 0.098 | CU467822.1 | -0.061 |
| slit1b | 0.074 | fabp7a | -0.061 |
| ndufa4 | 0.073 | atp1a1b | -0.058 |
| ywhag2 | 0.070 | mdkb | -0.056 |
| map1b | 0.070 | zgc:165461 | -0.053 |
| stx1b | 0.068 | glula | -0.049 |
| cplx2l | 0.068 | gpm6bb | -0.049 |
| zgc:65894 | 0.067 | mdka | -0.048 |
| stxbp1a | 0.066 | slc1a2b | -0.048 |
| si:ch73-119p20.1 | 0.066 | sox3 | -0.048 |
| atp1b2a | 0.066 | atp1b4 | -0.047 |
| calm2a | 0.066 | ppap2d | -0.044 |
| arxa | 0.065 | si:ch211-251b21.1 | -0.042 |
| nsfa | 0.065 | CR848047.1 | -0.041 |
| npr3 | 0.065 | slc6a11b | -0.040 |
| sypb | 0.065 | cx43 | -0.040 |
| hjv | 0.065 | her4.2 | -0.039 |
| clu | 0.064 | id1 | -0.039 |
| nptna | 0.064 | cspg5b | -0.038 |
| calm1a | 0.063 | msi1 | -0.038 |
| atp1a3a | 0.063 | her4.1 | -0.037 |
| stmn2a | 0.063 | eno1b | -0.036 |
| atp6v0cb | 0.063 | dap1b | -0.035 |
| syt1a | 0.063 | ptgdsb.2 | -0.035 |
| cers3b | 0.063 | si:ch73-30b17.2 | -0.035 |
| atp2b3a | 0.062 | XLOC-003690 | -0.035 |
| tubb2 | 0.062 | slc1a3b | -0.035 |
| map6b | 0.062 | notch3 | -0.035 |
| zgc:153426 | 0.062 | mfge8a | -0.035 |
| elavl4 | 0.062 | ptprz1b | -0.034 |
| si:ch211-214j24.9 | 0.062 | slc3a2a | -0.034 |
| vasna | 0.062 | her4.4 | -0.034 |
| gng3 | 0.062 | slc6a9 | -0.033 |
| lgals2a | 0.062 | CU634008.1 | -0.033 |
| atp6ap2 | 0.062 | gas1b | -0.033 |