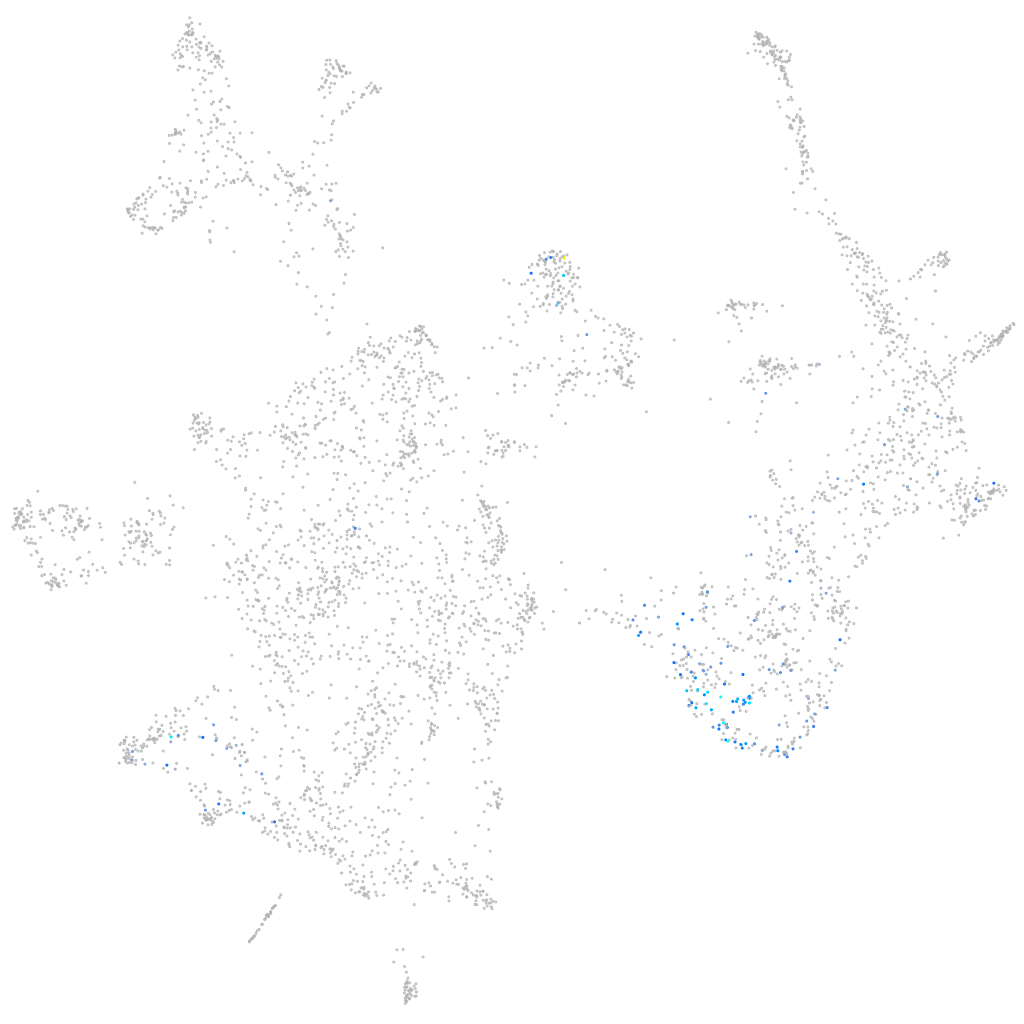
chromogranin B
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm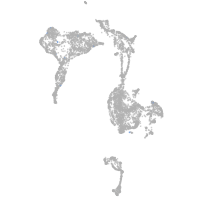 |
muscle 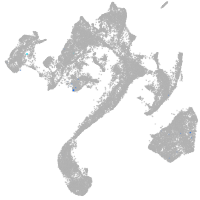 |
mural cells / non-skeletal muscle 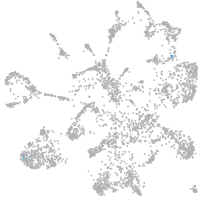 |
mesenchyme 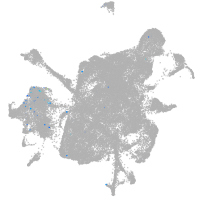 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory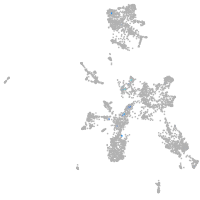 |
otic / lateral line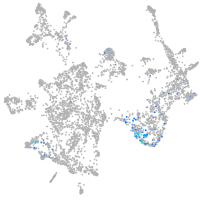 |
epidermis |
periderm |
fin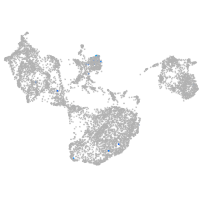 |
ionocytes / mucous |
pigment cells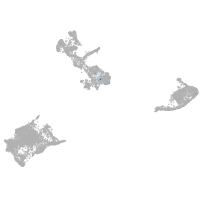 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| crip1 | 0.300 | si:ch211-243g18.2 | -0.117 |
| col17a1b | 0.271 | zgc:193541 | -0.115 |
| fat1b | 0.265 | stm | -0.107 |
| pcdh12 | 0.250 | calm2b | -0.102 |
| gdpd2 | 0.242 | atp5mc1 | -0.102 |
| CR388166.1 | 0.239 | cycsb | -0.094 |
| itgb6 | 0.229 | calm3b | -0.092 |
| LO017650.1 | 0.228 | snrpd1 | -0.091 |
| ankrd1a | 0.227 | sub1a | -0.090 |
| zte38 | 0.223 | mt-cyb | -0.089 |
| tnfrsf9a | 0.219 | cox8a | -0.088 |
| fermt1 | 0.215 | cox5ab | -0.088 |
| slc6a4b | 0.213 | cox7b | -0.087 |
| sfrp1a | 0.211 | psmb1 | -0.087 |
| igfbp5b | 0.204 | acbd7 | -0.087 |
| lmna | 0.204 | ptges3b | -0.086 |
| ptgis | 0.201 | otomp | -0.086 |
| si:dkey-33c9.6 | 0.200 | pax2a | -0.085 |
| LOC110439438 | 0.195 | sox10 | -0.085 |
| CT990561.1 | 0.193 | agr2 | -0.085 |
| tspan11 | 0.192 | hmgn3 | -0.084 |
| sostdc1a | 0.192 | tbx2b | -0.084 |
| myo10l1 | 0.191 | snrpg | -0.084 |
| tfap2a | 0.191 | atp5pd | -0.084 |
| frem2a | 0.187 | cfl1 | -0.084 |
| CRACR2A | 0.186 | psmb7 | -0.083 |
| lmo4b | 0.185 | copz1 | -0.083 |
| ackr3b | 0.185 | mt-nd1 | -0.083 |
| cryaba | 0.185 | snrpb | -0.082 |
| CT030188.1 | 0.185 | atp5if1a | -0.080 |
| nkx3.2 | 0.183 | atp5mc3a | -0.080 |
| syngap1a | 0.180 | cldna | -0.080 |
| celsr1b | 0.179 | eif5a2 | -0.080 |
| tagln2 | 0.178 | atp5fa1 | -0.079 |
| cyp8b2 | 0.177 | atp5f1b | -0.078 |