cofilin 1
ZFIN 
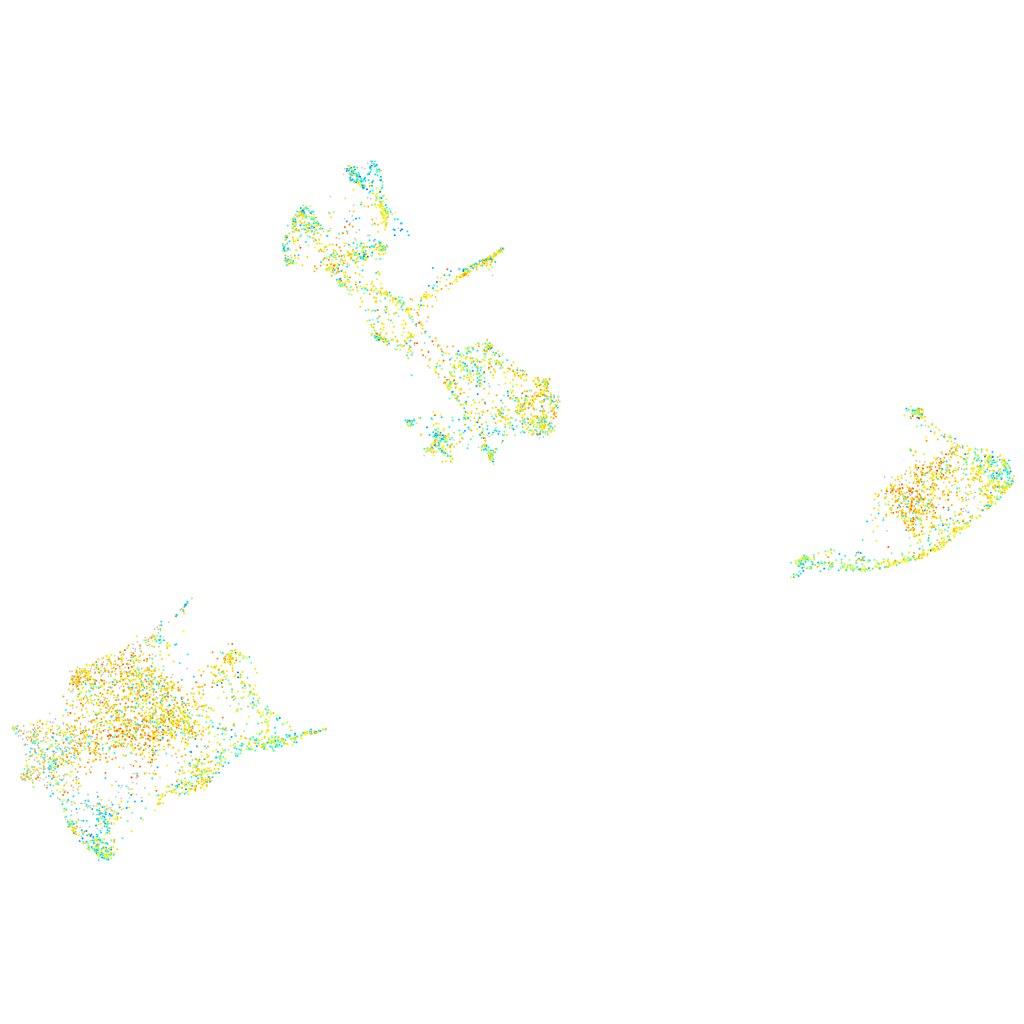
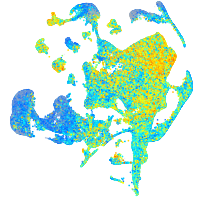
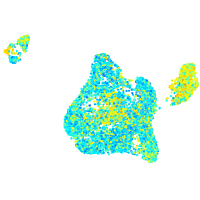
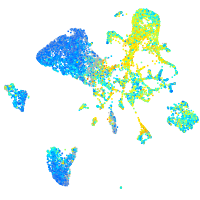
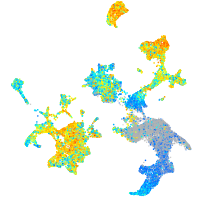
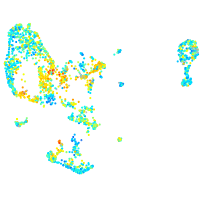
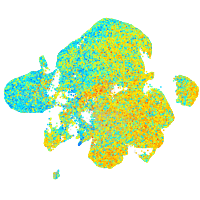
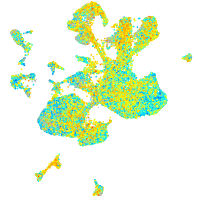
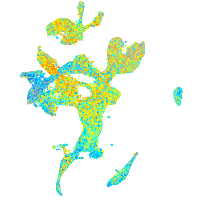
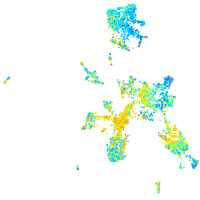
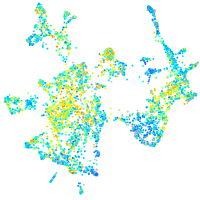
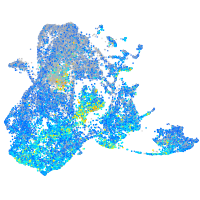
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ppiab | 0.190 | pvalb1 | -0.179 |
| psmb7 | 0.188 | apoa2 | -0.176 |
| qdpra | 0.184 | pvalb2 | -0.173 |
| atp5mc1 | 0.173 | apoa1b | -0.160 |
| calm2b | 0.172 | mylz3 | -0.159 |
| ywhabl | 0.169 | prss59.2 | -0.153 |
| abracl | 0.168 | actc1b | -0.150 |
| atp5f1b | 0.167 | CELA1 (1 of many) | -0.144 |
| tuba8l3 | 0.166 | prss1 | -0.143 |
| ptges3b | 0.166 | ctrb1 | -0.141 |
| cox7a2a | 0.166 | dio3a | -0.138 |
| atp5f1d | 0.165 | mylpfa | -0.122 |
| cox8a | 0.164 | sesn1 | -0.113 |
| slc25a3b | 0.163 | tnni2a.4 | -0.107 |
| atp6v1g1 | 0.162 | tnnt3b | -0.105 |
| cirbpa | 0.161 | ela2l | -0.103 |
| pfn2l | 0.161 | icn | -0.103 |
| rab1ab | 0.160 | krt4 | -0.102 |
| rpl17 | 0.160 | lpin1 | -0.100 |
| atp5pd | 0.160 | agmo | -0.100 |
| actb1 | 0.159 | nme2b.2 | -0.099 |
| arhgdia | 0.158 | krt17 | -0.099 |
| arpc2 | 0.156 | prss59.1 | -0.097 |
| tubb2b | 0.155 | zgc:112160 | -0.089 |
| vat1 | 0.154 | krt5 | -0.087 |
| prdx2 | 0.153 | cyt1 | -0.087 |
| mibp2 | 0.153 | aqp3a | -0.085 |
| prdx6 | 0.153 | myhz1.1 | -0.082 |
| psma5 | 0.153 | c6ast4 | -0.079 |
| atp6v1e1b | 0.152 | ela3l | -0.079 |
| cst14a.2 | 0.151 | sik1 | -0.078 |
| gdi2 | 0.151 | si:dkey-33c14.3 | -0.077 |
| psmb1 | 0.150 | fabp10a | -0.076 |
| uqcrfs1 | 0.149 | si:ch211-195b11.3 | -0.076 |
| cycsb | 0.149 | CR848833.1 | -0.076 |