cilia and flagella associated protein 43
ZFIN 
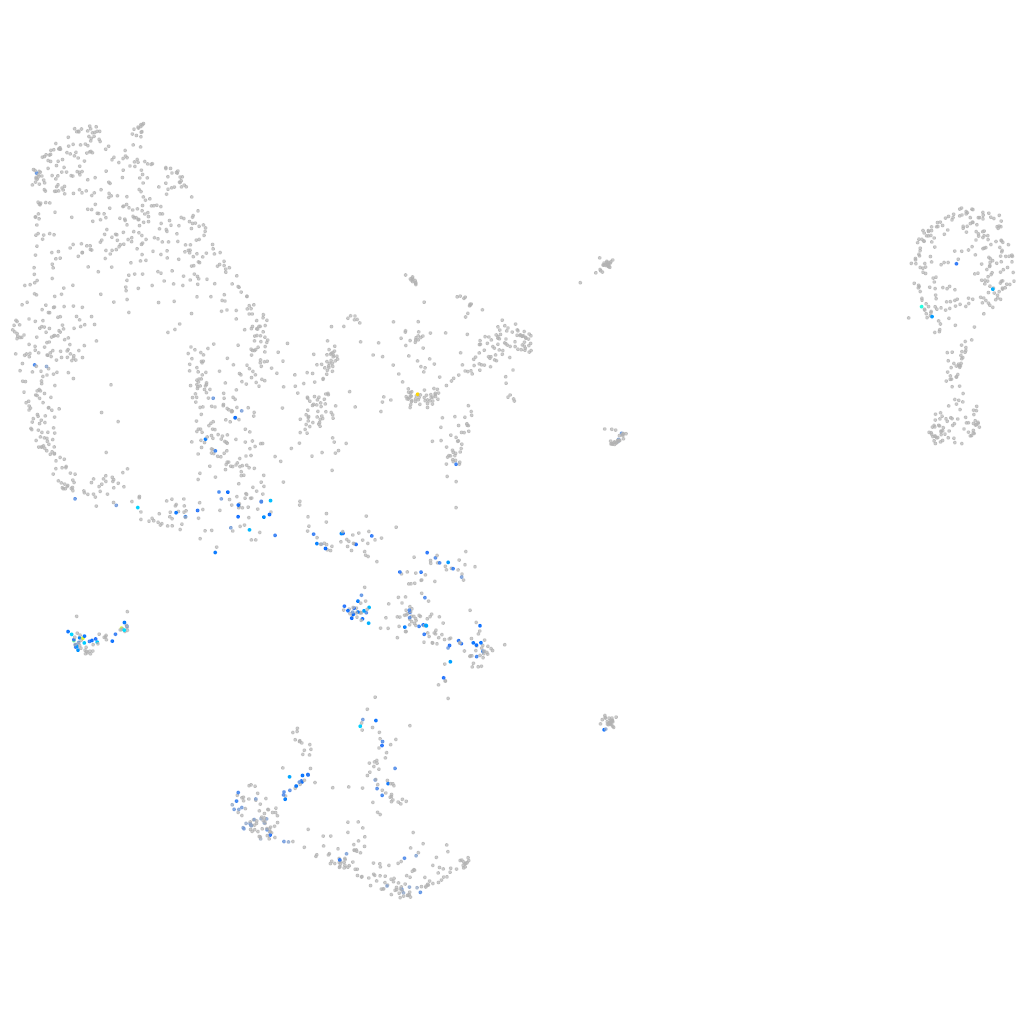
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm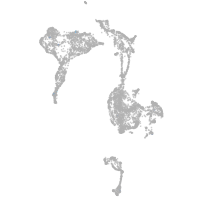 |
muscle 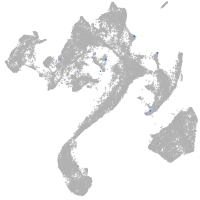 |
mural cells / non-skeletal muscle 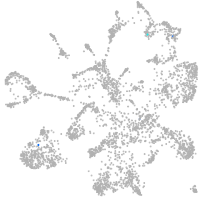 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 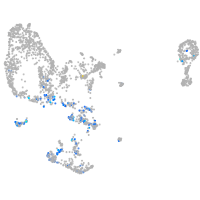 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| iqcg | 0.347 | gapdh | -0.158 |
| cfap45 | 0.343 | gcshb | -0.153 |
| ccdc114 | 0.334 | hao1 | -0.152 |
| capsla | 0.333 | aldob | -0.146 |
| si:ch211-163l21.7 | 0.331 | si:dkey-33i11.9 | -0.142 |
| zgc:55461 | 0.331 | fbp1b | -0.141 |
| enkur | 0.330 | gstt1a | -0.141 |
| si:dkey-148f10.4 | 0.327 | alpl | -0.140 |
| spag6 | 0.326 | cx32.3 | -0.139 |
| cfap58 | 0.325 | ASS1 | -0.138 |
| rsph9 | 0.324 | pnp6 | -0.138 |
| ccdc173 | 0.320 | dpys | -0.137 |
| dnali1 | 0.319 | agxtb | -0.135 |
| spaca9 | 0.318 | nipsnap3a | -0.133 |
| ropn1l | 0.318 | cubn | -0.133 |
| erich2 | 0.318 | epdl2 | -0.133 |
| pih1d3 | 0.316 | dab2 | -0.133 |
| catip | 0.316 | akr7a3 | -0.132 |
| ccdc151 | 0.315 | nit2 | -0.131 |
| cdc25d | 0.314 | gpt2l | -0.131 |
| LOC100329490 | 0.313 | lgmn | -0.129 |
| zgc:195356 | 0.311 | sod1 | -0.128 |
| tekt4 | 0.309 | gamt | -0.127 |
| tekt2 | 0.307 | phyhd1 | -0.126 |
| ccdc78 | 0.307 | grhprb | -0.126 |
| theg | 0.304 | smim1 | -0.125 |
| tekt1 | 0.304 | ctsz | -0.125 |
| cfap20 | 0.304 | sord | -0.125 |
| si:dkey-208b23.5 | 0.301 | g6pca.2 | -0.124 |
| ccdc40 | 0.300 | gstz1 | -0.124 |
| ift27 | 0.298 | rgn | -0.124 |
| spa17 | 0.298 | upb1 | -0.124 |
| cfap57 | 0.297 | aco1 | -0.124 |
| rsph4a | 0.297 | zgc:136493 | -0.123 |
| spata4 | 0.295 | slc20a1a | -0.123 |