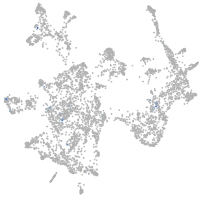
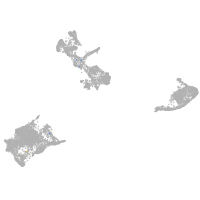
centrosomal protein 128
ZFIN 
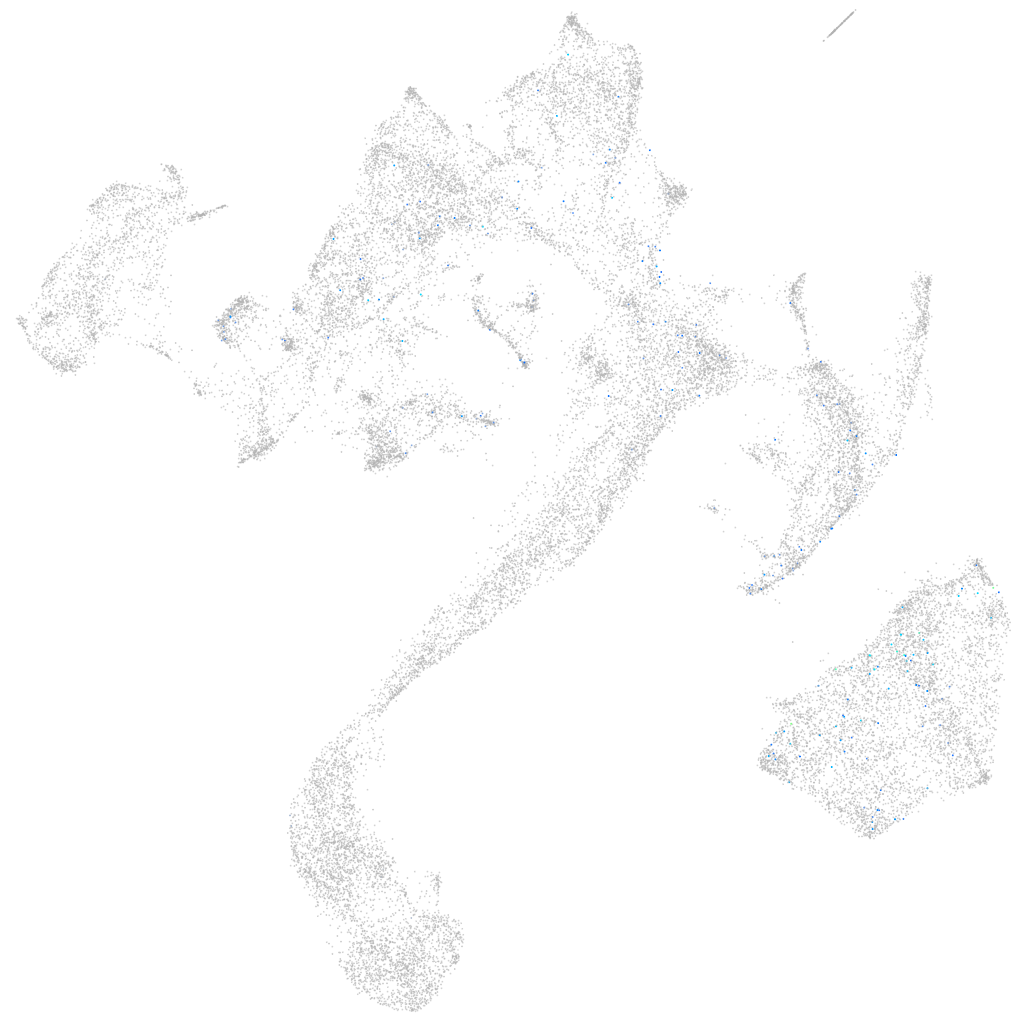
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX296552.1 | 0.140 | actc1b | -0.055 |
| XLOC-028452 | 0.099 | ak1 | -0.049 |
| cd79a | 0.092 | aldoab | -0.049 |
| clrn1 | 0.091 | vdac3 | -0.048 |
| LOC110438497 | 0.090 | rpl37 | -0.048 |
| XLOC-011738 | 0.081 | ttn.2 | -0.047 |
| LOC794378 | 0.080 | atp2a1 | -0.046 |
| mki67 | 0.079 | ckmb | -0.046 |
| mir723 | 0.078 | ckma | -0.046 |
| BX000364.1 | 0.077 | tmem38a | -0.046 |
| si:dkey-226n24.1 | 0.076 | ttn.1 | -0.046 |
| XLOC-011850 | 0.074 | zgc:114188 | -0.045 |
| ftr43 | 0.073 | tpi1b | -0.045 |
| CR376825.1 | 0.072 | tnnc2 | -0.045 |
| banf1 | 0.072 | acta1b | -0.044 |
| LOC103910514 | 0.071 | desma | -0.044 |
| XLOC-002642 | 0.071 | tpma | -0.044 |
| seta | 0.069 | rps10 | -0.044 |
| supt16h | 0.069 | mybphb | -0.044 |
| mir10a | 0.069 | ldb3a | -0.044 |
| smc2 | 0.068 | acta1a | -0.044 |
| CU466285.1 | 0.068 | srl | -0.044 |
| rrm1 | 0.068 | klhl41b | -0.044 |
| zgc:110425 | 0.068 | eno1a | -0.043 |
| si:ch211-137j23.6 | 0.067 | romo1 | -0.043 |
| tubb2b | 0.067 | vdac2 | -0.043 |
| lig1 | 0.067 | atp5if1b | -0.043 |
| hmgb2a | 0.067 | rps17 | -0.043 |
| stmn1a | 0.066 | cav3 | -0.042 |
| anp32a | 0.066 | atp5meb | -0.042 |
| tpx2 | 0.066 | neb | -0.042 |
| ptmab | 0.066 | CABZ01078594.1 | -0.042 |
| dek | 0.066 | mylpfa | -0.042 |
| baz1b | 0.066 | gapdh | -0.041 |
| chaf1a | 0.065 | txlnbb | -0.041 |