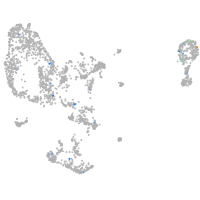
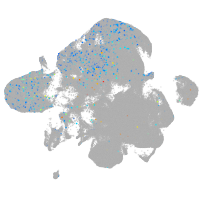
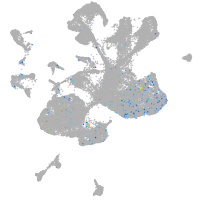
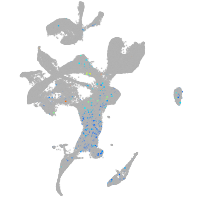
centromere protein Q
ZFIN 
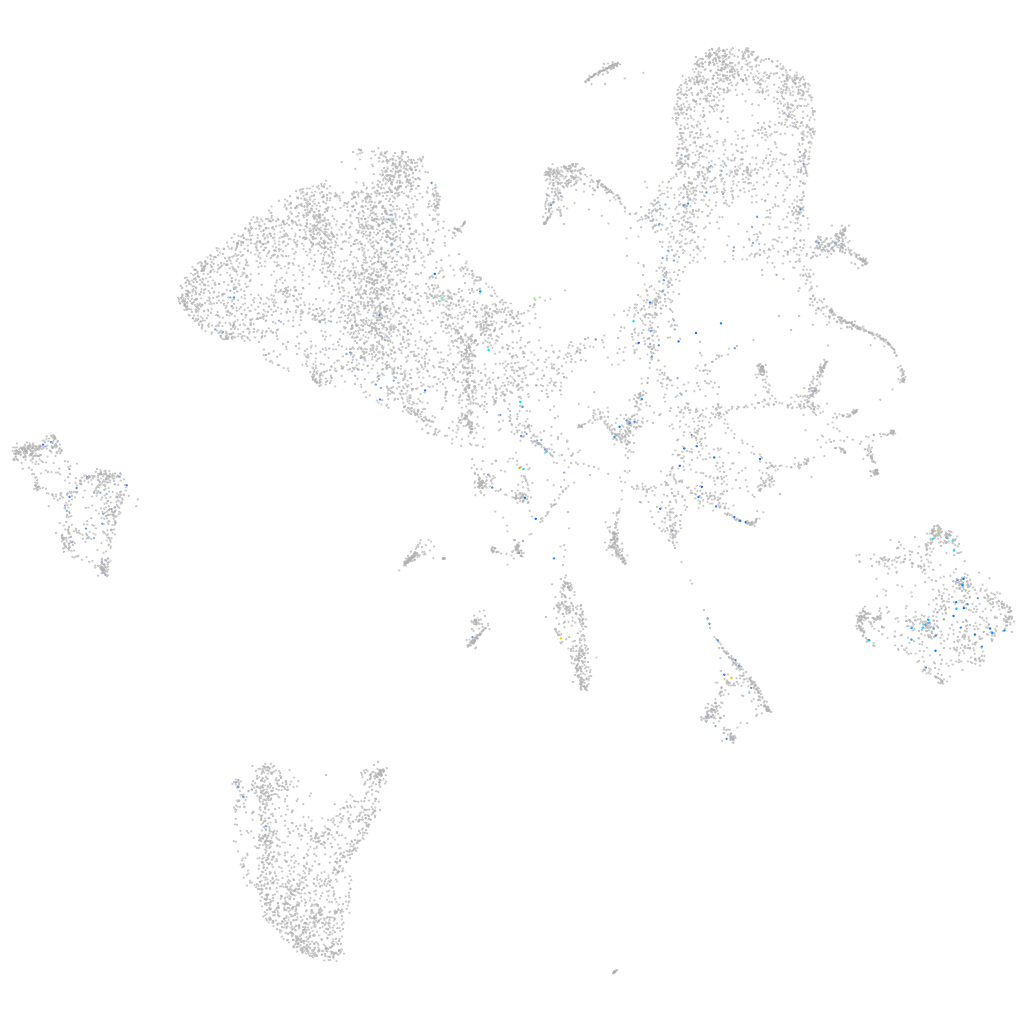
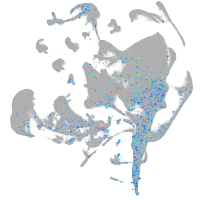
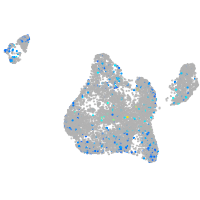
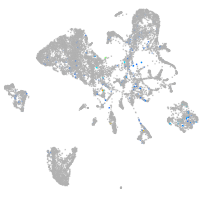
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tunar | 0.258 | gatm | -0.069 |
| LOC101884089 | 0.176 | gamt | -0.067 |
| si:dkey-261m9.9 | 0.172 | gapdh | -0.062 |
| mmp28 | 0.171 | pnp4b | -0.062 |
| pcna | 0.145 | zgc:92744 | -0.061 |
| mibp | 0.145 | cx32.3 | -0.057 |
| zgc:110540 | 0.144 | dap | -0.057 |
| rbbp4 | 0.138 | glud1b | -0.056 |
| atad5a | 0.137 | gpx4a | -0.056 |
| stmn1a | 0.137 | abat | -0.055 |
| anp32b | 0.136 | g6pca.2 | -0.054 |
| XLOC-015238 | 0.136 | ahcy | -0.054 |
| lig1 | 0.133 | agxtb | -0.054 |
| dnmt1 | 0.132 | ckba | -0.054 |
| cenpn | 0.130 | mat1a | -0.053 |
| dut | 0.130 | phyh | -0.053 |
| esco2 | 0.128 | rbp4 | -0.053 |
| nasp | 0.128 | aqp12 | -0.052 |
| rrm1 | 0.128 | fetub | -0.052 |
| LOC101882375 | 0.128 | pklr | -0.052 |
| tuba8l4 | 0.127 | gda | -0.052 |
| nsd2 | 0.126 | bhmt | -0.052 |
| dek | 0.126 | apobb.1 | -0.051 |
| dhfr | 0.125 | pck1 | -0.051 |
| slbp | 0.124 | pvalb1 | -0.050 |
| seta | 0.124 | ppdpfa | -0.050 |
| tubb2b | 0.124 | ces2 | -0.050 |
| chaf1a | 0.124 | apoa4b.1 | -0.050 |
| lbr | 0.124 | eif4ebp3 | -0.050 |
| hmgb2a | 0.123 | grhprb | -0.049 |
| CABZ01062796.1 | 0.123 | c9 | -0.049 |
| cbx3a | 0.122 | ndrg2 | -0.049 |
| nucks1b | 0.121 | LOC110437731 | -0.049 |
| BX682558.1 | 0.121 | cyp2ad2 | -0.049 |
| rfc3 | 0.120 | rps10 | -0.049 |