centromere protein N
ZFIN 
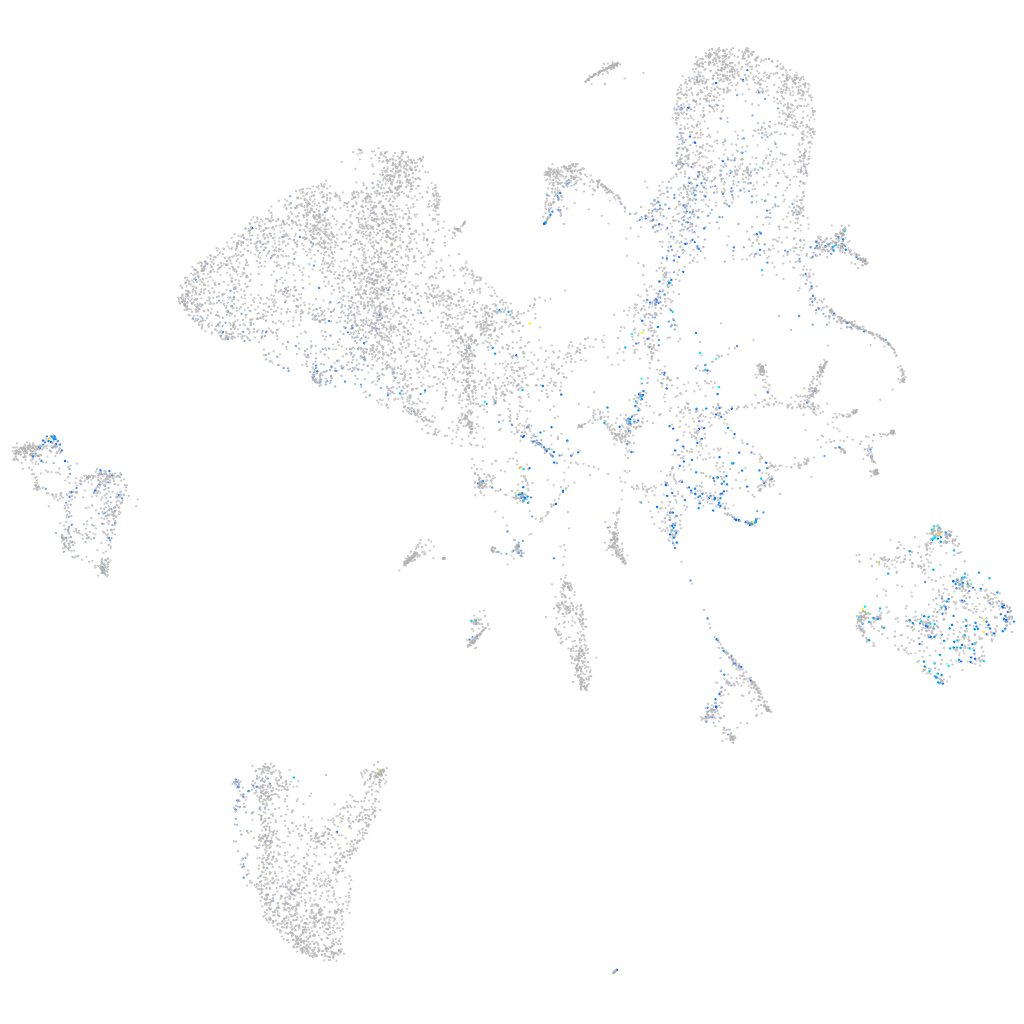
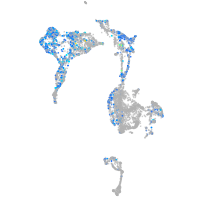
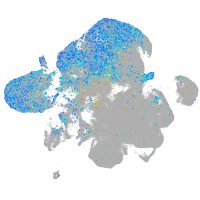
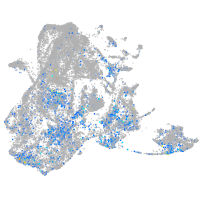
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccna2 | 0.331 | gamt | -0.178 |
| pcna | 0.329 | gatm | -0.163 |
| stmn1a | 0.324 | gapdh | -0.155 |
| mki67 | 0.324 | bhmt | -0.153 |
| tubb2b | 0.322 | pnp4b | -0.146 |
| rrm1 | 0.319 | gpx4a | -0.143 |
| cks1b | 0.317 | zgc:92744 | -0.140 |
| hmgb2a | 0.316 | apoa2 | -0.140 |
| hmgb2b | 0.310 | pklr | -0.139 |
| anp32b | 0.300 | apoa1b | -0.139 |
| seta | 0.298 | apoa4b.1 | -0.134 |
| cbx3a | 0.298 | rbp4 | -0.133 |
| tuba8l4 | 0.297 | agxtb | -0.133 |
| aurkb | 0.296 | mat1a | -0.131 |
| dek | 0.295 | nupr1b | -0.131 |
| cdk1 | 0.294 | fetub | -0.127 |
| hmga1a | 0.291 | abat | -0.127 |
| dut | 0.289 | afp4 | -0.127 |
| chaf1a | 0.286 | zgc:123103 | -0.126 |
| rpa2 | 0.285 | ces2 | -0.125 |
| lbr | 0.285 | aldob | -0.124 |
| mad2l1 | 0.283 | fbp1b | -0.123 |
| ncapd2 | 0.283 | ckba | -0.123 |
| smc2 | 0.283 | grhprb | -0.123 |
| banf1 | 0.283 | apobb.1 | -0.123 |
| dlgap5 | 0.281 | aqp12 | -0.122 |
| sumo3b | 0.280 | apom | -0.122 |
| si:ch211-288g17.3 | 0.280 | fabp10a | -0.122 |
| mibp | 0.279 | g6pca.2 | -0.122 |
| tpx2 | 0.277 | glud1b | -0.121 |
| nasp | 0.276 | serpina1 | -0.121 |
| lig1 | 0.274 | LOC110437731 | -0.120 |
| si:ch73-281n10.2 | 0.273 | tfa | -0.119 |
| si:dkey-6i22.5 | 0.273 | mgst1.2 | -0.119 |
| rrm2 | 0.273 | ahcy | -0.119 |